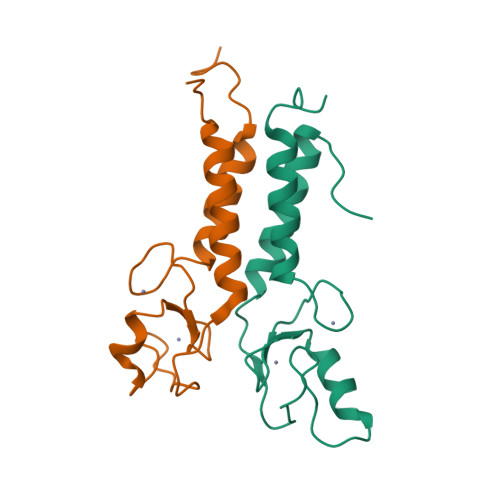
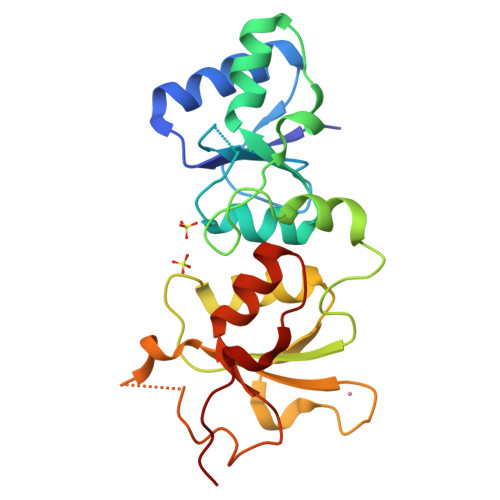
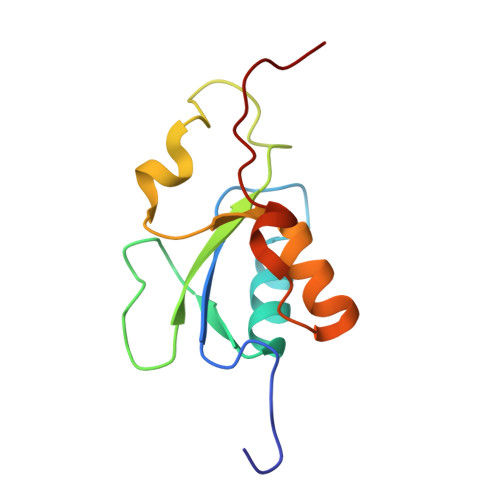
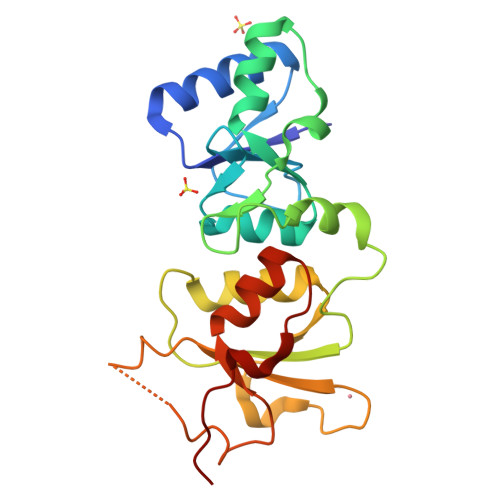
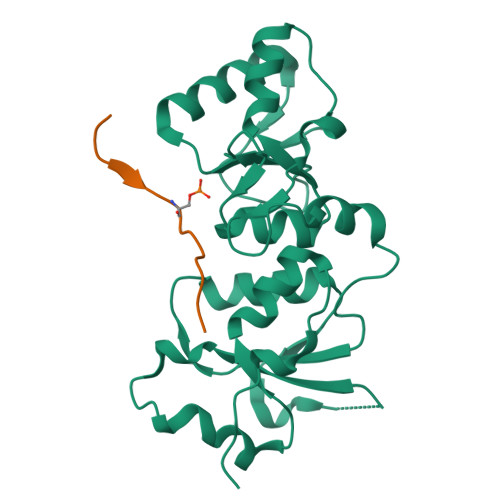
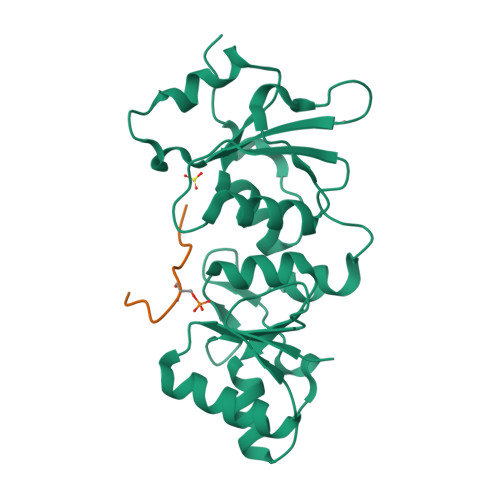
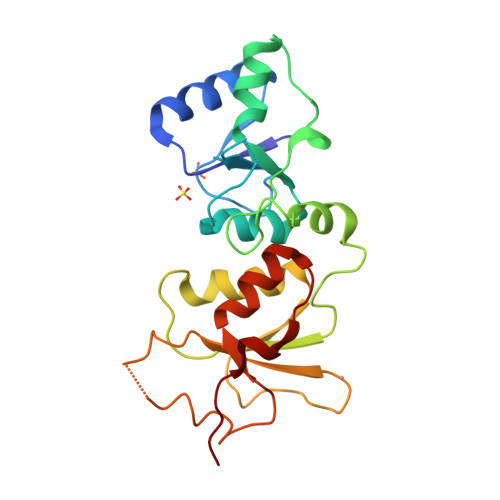
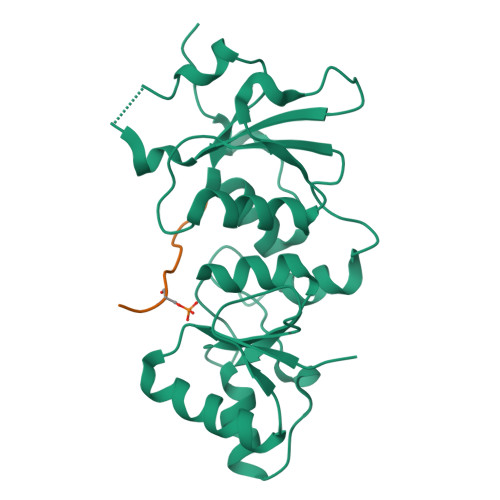
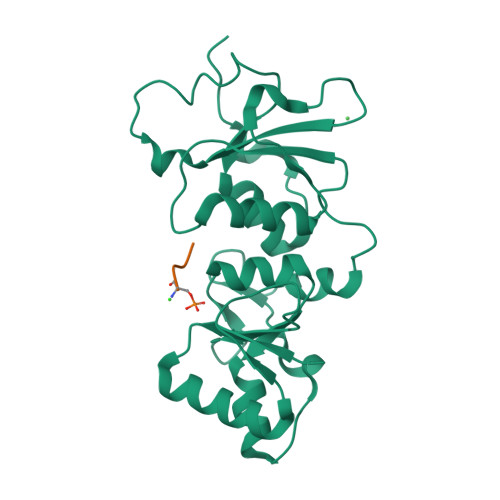
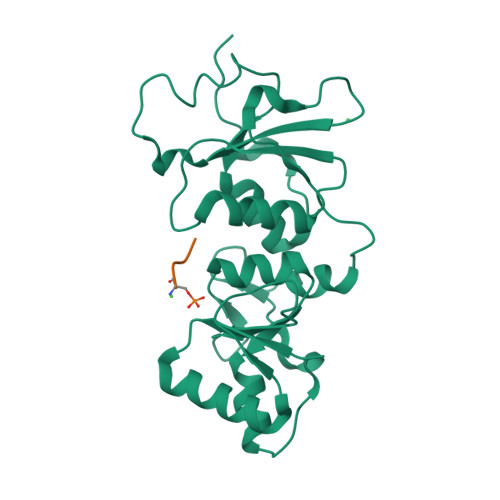
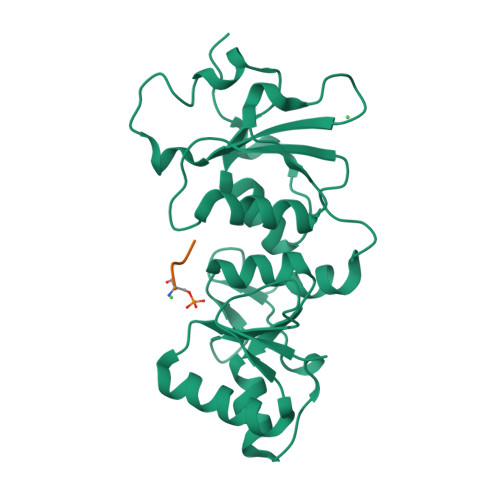
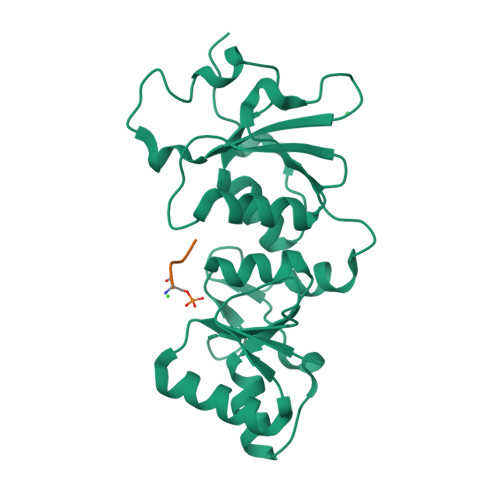
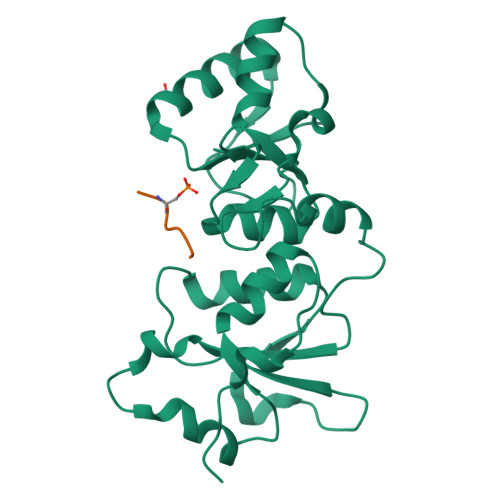
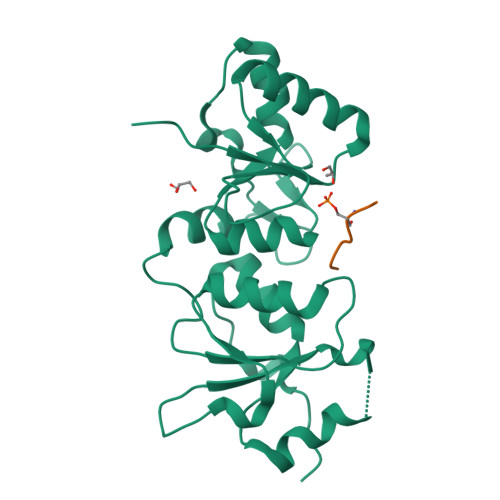
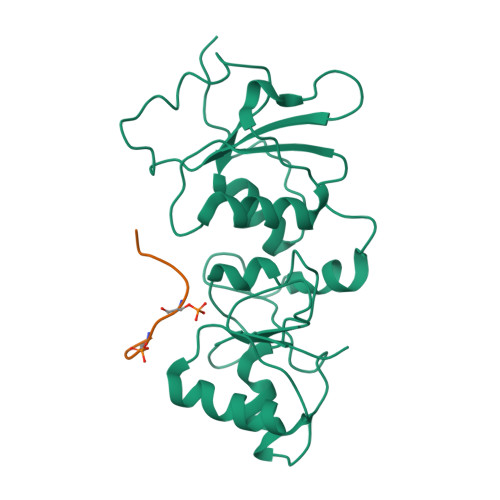
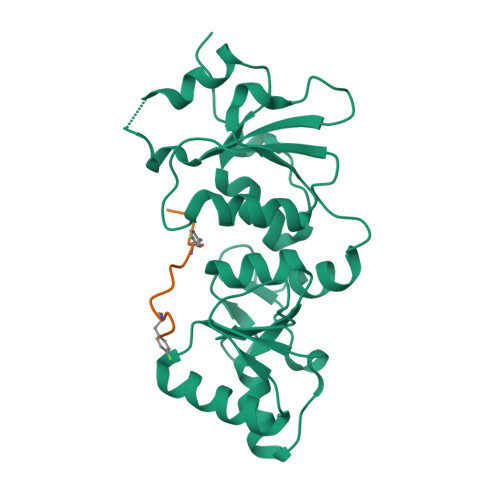
| # ALTERNATIVE PRODUCTS | BRCA1_HUMAN | Event=Alternative splicing, Alternative initiation; Named isoforms=8; Name=1; IsoId=P38398-1; Sequence=Displayed; Name=2; IsoId=P38398-2; Sequence=VSP_047891; Note=May be produced at very low levels due to a premature stop codon in the mRNA, leading to nonsense-mediated mRNA decay.; Name=3; Synonyms=Delta11b; IsoId=P38398-3; Sequence=VSP_035399, VSP_043797; Name=4; Synonyms=DeltaBRCA1(17aa); IsoId=P38398-4; Sequence=VSP_035396; Note=Produced by alternative initiation at Met-18 of isoform 1.; Name=5; Synonyms=Delta11, Delta772-3095; IsoId=P38398-5; Sequence=VSP_035398; Name=6; IsoId=P38398-6; Sequence=VSP_035399, VSP_043797, VSP_043798; Note=No experimental confirmation available.; Name=7; IsoId=P38398-7; Sequence=VSP_055404; Note=No experimental confirmation available. Ref.8 (AAI15038) sequence is in conflict in position 1461 N->D. {ECO 0000305}; Name=8; IsoId=P38398-8; Sequence=VSP_057569; Note=No experimental confirmation available. The N-terminus is confirmed by several cDNAs. {ECO 0000305}; |
| # AltName | BRCA1_HUMAN | RING finger protein 53 |
| # AltName | RING-type E3 ubiquitin transferase BRCA1 {ECO | 0000305} |
| # BRENDA | 6.3.2.19 | 2681 |
| # BioGrid | 107140 | 566 |
| # CATALYTIC ACTIVITY | BRCA1_HUMAN | S-ubiquitinyl-[E2 ubiquitin-conjugating enzyme]-L-cysteine + [acceptor protein]-L-lysine = [E2 ubiquitin- conjugating enzyme]-L-cysteine + N(6)-ubiquitinyl-[acceptor protein]-L-lysine. {ECO 0000269|PubMed 10500182, ECO 0000269|PubMed 12887909, ECO 0000269|PubMed 12890688, ECO 0000269|PubMed 16818604, ECO 0000269|PubMed 18056443, ECO 0000269|PubMed 20351172}. |
| # CCDS | CCDS11453 | -. [P38398-1] |
| # CCDS | CCDS11454 | -. [P38398-3] |
| # CCDS | CCDS11455 | -. [P38398-6] |
| # CCDS | CCDS11456 | -. [P38398-7] |
| # CCDS | CCDS11459 | -. [P38398-8] |
| # CDD | cd00027 | BRCT; 2 |
| # ChiTaRS | BRCA1 | human |
| # DISEASE | BRCA1_HUMAN | Breast cancer (BC) [MIM 114480] A common malignancy originating from breast epithelial tissue. Breast neoplasms can be distinguished by their histologic pattern. Invasive ductal carcinoma is by far the most common type. Breast cancer is etiologically and genetically heterogeneous. Important genetic factors have been indicated by familial occurrence and bilateral involvement. Mutations at more than one locus can be involved in different families or even in the same case. {ECO 0000269|PubMed 10323242, ECO 0000269|PubMed 12442275, ECO 0000269|PubMed 12938098, ECO 0000269|PubMed 14722926, ECO 0000269|PubMed 18285836, ECO 0000269|PubMed 7545954, ECO 0000269|PubMed 7894491, ECO 0000269|PubMed 7894493, ECO 0000269|PubMed 7939630, ECO 0000269|PubMed 8554067, ECO 0000269|PubMed 8723683, ECO 0000269|PubMed 8776600, ECO 0000269|PubMed 9482581, ECO 0000269|PubMed 9609997, ECO 0000269|PubMed 9760198}. Note=Disease susceptibility is associated with variations affecting the gene represented in this entry. Mutations in BRCA1 are thought to be responsible for 45% of inherited breast cancer. Moreover, BRCA1 carriers have a 4-fold increased risk of colon cancer, whereas male carriers face a 3- fold increased risk of prostate cancer. Cells lacking BRCA1 show defects in DNA repair by homologous recombination. |
| # DISEASE | BRCA1_HUMAN | Breast-ovarian cancer, familial, 1 (BROVCA1) [MIM 604370] A condition associated with familial predisposition to cancer of the breast and ovaries. Characteristic features in affected families are an early age of onset of breast cancer (often before age 50), increased chance of bilateral cancers (cancer that develop in both breasts, or both ovaries, independently), frequent occurrence of breast cancer among men, increased incidence of tumors of other specific organs, such as the prostate. {ECO 0000269|PubMed 12938098, ECO 0000269|PubMed 14722926, ECO 0000269|PubMed 8968716}. Note=Disease susceptibility is associated with variations affecting the gene represented in this entry. Mutations in BRCA1 are thought to be responsible for more than 80% of inherited breast-ovarian cancer. |
| # DISEASE | BRCA1_HUMAN | Ovarian cancer (OC) [MIM 167000] The term ovarian cancer defines malignancies originating from ovarian tissue. Although many histologic types of ovarian tumors have been described, epithelial ovarian carcinoma is the most common form. Ovarian cancers are often asymptomatic and the recognized signs and symptoms, even of late-stage disease, are vague. Consequently, most patients are diagnosed with advanced disease. {ECO 0000269|PubMed 10196379, ECO 0000269|PubMed 10486320, ECO 0000269|PubMed 14746861}. Note=Disease susceptibility is associated with variations affecting the gene represented in this entry. |
| # DISEASE | BRCA1_HUMAN | Pancreatic cancer 4 (PNCA4) [MIM 614320] A malignant neoplasm of the pancreas. Tumors can arise from both the exocrine and endocrine portions of the pancreas, but 95% of them develop from the exocrine portion, including the ductal epithelium, acinar cells, connective tissue, and lymphatic tissue. {ECO 0000269|PubMed 18762988}. Note=Disease susceptibility is associated with variations affecting the gene represented in this entry. |
| # DOMAIN | BRCA1_HUMAN | The BRCT domains recognize and bind phosphorylated pSXXF motif on proteins. The interaction with the phosphorylated pSXXF motif of FAM175A/Abraxas, recruits BRCA1 at DNA damage sites. {ECO 0000269|PubMed 20159462}. |
| # DOMAIN | BRCA1_HUMAN | The RING-type zinc finger domain interacts with BAP1. {ECO 0000269|PubMed 20159462}. |
| # ENZYME REGULATION | The E3 ubiquitin-protein ligase activity is inhibited by phosphorylation by AURKA. Activity is increased by phosphatase treatment. {ECO:0000269|PubMed | 18056443}. |
| # Ensembl | ENST00000352993 | ENSP00000312236; ENSG00000012048. [P38398-5] |
| # Ensembl | ENST00000357654 | ENSP00000350283; ENSG00000012048. [P38398-1] |
| # Ensembl | ENST00000461221 | ENSP00000418548; ENSG00000012048. [P38398-2] |
| # Ensembl | ENST00000461798 | ENSP00000417988; ENSG00000012048. [P38398-2] |
| # Ensembl | ENST00000468300 | ENSP00000417148; ENSG00000012048. [P38398-6] |
| # Ensembl | ENST00000471181 | ENSP00000418960; ENSG00000012048. [P38398-7] |
| # Ensembl | ENST00000491747 | ENSP00000420705; ENSG00000012048. [P38398-3] |
| # Ensembl | ENST00000493795 | ENSP00000418775; ENSG00000012048. [P38398-8] |
| # ExpressionAtlas | P38398 | baseline and differential |
| # FUNCTION | BRCA1_HUMAN | E3 ubiquitin-protein ligase that specifically mediates the formation of 'Lys-6'-linked polyubiquitin chains and plays a central role in DNA repair by facilitating cellular responses to DNA damage. It is unclear whether it also mediates the formation of other types of polyubiquitin chains. The E3 ubiquitin-protein ligase activity is required for its tumor suppressor function. The BRCA1-BARD1 heterodimer coordinates a diverse range of cellular pathways such as DNA damage repair, ubiquitination and transcriptional regulation to maintain genomic stability. Regulates centrosomal microtubule nucleation. Required for normal cell cycle progression from G2 to mitosis. Required for appropriate cell cycle arrests after ionizing irradiation in both the S-phase and the G2 phase of the cell cycle. Involved in transcriptional regulation of P21 in response to DNA damage. Required for FANCD2 targeting to sites of DNA damage. May function as a transcriptional regulator. Inhibits lipid synthesis by binding to inactive phosphorylated ACACA and preventing its dephosphorylation. Contributes to homologous recombination repair (HRR) via its direct interaction with PALB2, fine-tunes recombinational repair partly through its modulatory role in the PALB2-dependent loading of BRCA2-RAD51 repair machinery at DNA breaks. Component of the BRCA1-RBBP8 complex which regulates CHEK1 activation and controls cell cycle G2/M checkpoints on DNA damage via BRCA1-mediated ubiquitination of RBBP8. Acts as a transcriptional activator (PubMed 20160719). {ECO 0000269|PubMed 10500182, ECO 0000269|PubMed 10724175, ECO 0000269|PubMed 11836499, ECO 0000269|PubMed 12887909, ECO 0000269|PubMed 12890688, ECO 0000269|PubMed 14976165, ECO 0000269|PubMed 14990569, ECO 0000269|PubMed 16326698, ECO 0000269|PubMed 16818604, ECO 0000269|PubMed 17525340, ECO 0000269|PubMed 18056443, ECO 0000269|PubMed 19261748, ECO 0000269|PubMed 19369211, ECO 0000269|PubMed 20160719, ECO 0000269|PubMed 20351172, ECO 0000269|PubMed 20364141}. |
| # GO_component | GO:0000151 | ubiquitin ligase complex; NAS:UniProtKB. |
| # GO_component | GO:0000800 | lateral element; IDA:MGI. |
| # GO_component | GO:0005634 | nucleus; IDA:UniProtKB. |
| # GO_component | GO:0005654 | nucleoplasm; TAS:Reactome. |
| # GO_component | GO:0005694 | chromosome; ISS:UniProtKB. |
| # GO_component | GO:0005737 | cytoplasm; IDA:UniProtKB. |
| # GO_component | GO:0005886 | plasma membrane; IDA:BHF-UCL. |
| # GO_component | GO:0008274 | gamma-tubulin ring complex; NAS:UniProtKB. |
| # GO_component | GO:0030529 | intracellular ribonucleoprotein complex; IDA:MGI. |
| # GO_component | GO:0031436 | BRCA1-BARD1 complex; IDA:UniProtKB. |
| # GO_component | GO:0043234 | protein complex; IDA:UniProtKB. |
| # GO_component | GO:0070531 | BRCA1-A complex; IDA:UniProtKB. |
| # GO_function | GO:0003677 | DNA binding; TAS:ProtInc. |
| # GO_function | GO:0003684 | damaged DNA binding; IEA:Ensembl. |
| # GO_function | GO:0003713 | transcription coactivator activity; NAS:UniProtKB. |
| # GO_function | GO:0003723 | RNA binding; IDA:MGI. |
| # GO_function | GO:0004842 | ubiquitin-protein transferase activity; IDA:UniProtKB. |
| # GO_function | GO:0008270 | zinc ion binding; TAS:ProtInc. |
| # GO_function | GO:0015631 | tubulin binding; NAS:UniProtKB. |
| # GO_function | GO:0019899 | enzyme binding; IPI:UniProtKB. |
| # GO_function | GO:0031625 | ubiquitin protein ligase binding; IPI:UniProtKB. |
| # GO_function | GO:0044212 | transcription regulatory region DNA binding; IDA:BHF-UCL. |
| # GO_function | GO:0050681 | androgen receptor binding; NAS:UniProtKB. |
| # GO_process | GO:0000724 | double-strand break repair via homologous recombination; IDA:HGNC. |
| # GO_process | GO:0000729 | DNA double-strand break processing; TAS:Reactome. |
| # GO_process | GO:0000731 | DNA synthesis involved in DNA repair; TAS:Reactome. |
| # GO_process | GO:0000732 | strand displacement; TAS:Reactome. |
| # GO_process | GO:0006260 | DNA replication; TAS:Reactome. |
| # GO_process | GO:0006301 | postreplication repair; IDA:HGNC. |
| # GO_process | GO:0006302 | double-strand break repair; IMP:UniProtKB. |
| # GO_process | GO:0006303 | double-strand break repair via nonhomologous end joining; TAS:Reactome. |
| # GO_process | GO:0006349 | regulation of gene expression by genetic imprinting; IEA:Ensembl. |
| # GO_process | GO:0006351 | transcription, DNA-templated; IEA:UniProtKB-KW. |
| # GO_process | GO:0006357 | regulation of transcription from RNA polymerase II promoter; TAS:ProtInc. |
| # GO_process | GO:0006359 | regulation of transcription from RNA polymerase III promoter; TAS:UniProtKB. |
| # GO_process | GO:0006633 | fatty acid biosynthetic process; IEA:UniProtKB-KW. |
| # GO_process | GO:0006915 | apoptotic process; TAS:UniProtKB. |
| # GO_process | GO:0006974 | cellular response to DNA damage stimulus; TAS:ProtInc. |
| # GO_process | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator; TAS:UniProtKB. |
| # GO_process | GO:0007059 | chromosome segregation; IMP:UniProtKB. |
| # GO_process | GO:0007098 | centrosome cycle; IEA:Ensembl. |
| # GO_process | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage; IDA:MGI. |
| # GO_process | GO:0009048 | dosage compensation by inactivation of X chromosome; IBA:GO_Central. |
| # GO_process | GO:0010212 | response to ionizing radiation; IMP:UniProtKB. |
| # GO_process | GO:0010575 | positive regulation of vascular endothelial growth factor production; IMP:BHF-UCL. |
| # GO_process | GO:0010628 | positive regulation of gene expression; IMP:BHF-UCL. |
| # GO_process | GO:0016567 | protein ubiquitination; IDA:HGNC. |
| # GO_process | GO:0016925 | protein sumoylation; TAS:Reactome. |
| # GO_process | GO:0030521 | androgen receptor signaling pathway; NAS:UniProtKB. |
| # GO_process | GO:0031052 | chromosome breakage; IEA:Ensembl. |
| # GO_process | GO:0031398 | positive regulation of protein ubiquitination; IDA:UniProtKB. |
| # GO_process | GO:0031572 | G2 DNA damage checkpoint; IMP:UniProtKB. |
| # GO_process | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway; IMP:CACAO. |
| # GO_process | GO:0035066 | positive regulation of histone acetylation; IDA:BHF-UCL. |
| # GO_process | GO:0035067 | negative regulation of histone acetylation; IBA:GO_Central. |
| # GO_process | GO:0042127 | regulation of cell proliferation; TAS:UniProtKB. |
| # GO_process | GO:0042981 | regulation of apoptotic process; TAS:UniProtKB. |
| # GO_process | GO:0043009 | chordate embryonic development; IBA:GO_Central. |
| # GO_process | GO:0043627 | response to estrogen; IDA:UniProtKB. |
| # GO_process | GO:0044030 | regulation of DNA methylation; IEA:Ensembl. |
| # GO_process | GO:0045717 | negative regulation of fatty acid biosynthetic process; IMP:UniProtKB. |
| # GO_process | GO:0045739 | positive regulation of DNA repair; IMP:UniProtKB. |
| # GO_process | GO:0045766 | positive regulation of angiogenesis; IMP:BHF-UCL. |
| # GO_process | GO:0045892 | negative regulation of transcription, DNA-templated; IDA:UniProtKB. |
| # GO_process | GO:0045893 | positive regulation of transcription, DNA-templated; IDA:UniProtKB. |
| # GO_process | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter; IDA:UniProtKB. |
| # GO_process | GO:0046600 | negative regulation of centriole replication; NAS:UniProtKB. |
| # GO_process | GO:0051571 | positive regulation of histone H3-K4 methylation; IDA:BHF-UCL. |
| # GO_process | GO:0051572 | negative regulation of histone H3-K4 methylation; IEA:Ensembl. |
| # GO_process | GO:0051573 | negative regulation of histone H3-K9 methylation; IDA:BHF-UCL. |
| # GO_process | GO:0051574 | positive regulation of histone H3-K9 methylation; IEA:Ensembl. |
| # GO_process | GO:0051865 | protein autoubiquitination; IDA:UniProtKB. |
| # GO_process | GO:0070512 | positive regulation of histone H4-K20 methylation; IDA:BHF-UCL. |
| # GO_process | GO:0071158 | positive regulation of cell cycle arrest; IDA:BHF-UCL. |
| # GO_process | GO:0071356 | cellular response to tumor necrosis factor; IMP:BHF-UCL. |
| # GO_process | GO:0071681 | cellular response to indole-3-methanol; IDA:UniProtKB. |
| # GO_process | GO:0085020 | protein K6-linked ubiquitination; IDA:UniProtKB. |
| # GO_process | GO:1901796 | regulation of signal transduction by p53 class mediator; TAS:Reactome. |
| # GO_process | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors; IMP:BHF-UCL. |
| # GO_process | GO:2000378 | negative regulation of reactive oxygen species metabolic process; IMP:BHF-UCL. |
| # GO_process | GO:2000617 | positive regulation of histone H3-K9 acetylation; IDA:BHF-UCL. |
| # GO_process | GO:2000620 | positive regulation of histone H4-K16 acetylation; IDA:BHF-UCL. |
| # GOslim_component | GO:0005575 | cellular_component |
| # GOslim_component | GO:0005634 | nucleus |
| # GOslim_component | GO:0005654 | nucleoplasm |
| # GOslim_component | GO:0005694 | chromosome |
| # GOslim_component | GO:0005737 | cytoplasm |
| # GOslim_component | GO:0005886 | plasma membrane |
| # GOslim_component | GO:0043234 | protein complex |
| # GOslim_function | GO:0000988 | transcription factor activity, protein binding |
| # GOslim_function | GO:0003674 | molecular_function |
| # GOslim_function | GO:0003677 | DNA binding |
| # GOslim_function | GO:0003723 | RNA binding |
| # GOslim_function | GO:0008092 | cytoskeletal protein binding |
| # GOslim_function | GO:0019899 | enzyme binding |
| # GOslim_function | GO:0043167 | ion binding |
| # GOslim_process | GO:0006259 | DNA metabolic process |
| # GOslim_process | GO:0006464 | cellular protein modification process |
| # GOslim_process | GO:0006629 | lipid metabolic process |
| # GOslim_process | GO:0006950 | response to stress |
| # GOslim_process | GO:0007059 | chromosome segregation |
| # GOslim_process | GO:0007165 | signal transduction |
| # GOslim_process | GO:0008150 | biological_process |
| # GOslim_process | GO:0008219 | cell death |
| # GOslim_process | GO:0009058 | biosynthetic process |
| # GOslim_process | GO:0009790 | embryo development |
| # GOslim_process | GO:0034641 | cellular nitrogen compound metabolic process |
| # GOslim_process | GO:0044281 | small molecule metabolic process |
| # GOslim_process | GO:0051276 | chromosome organization |
| # Gene3D | 3.30.40.10 | -; 1. |
| # Gene3D | 3.40.50.10190 | -; 2. |
| # Genevisible | P38398 | HS |
| # HGNC | HGNC:1100 | BRCA1 |
| # INTERACTION | BRCA1_HUMAN | Q13085 ACACA; NbExp=2; IntAct=EBI-349905, EBI-717681; Q92560 BAP1; NbExp=3; IntAct=EBI-349905, EBI-1791447; Q99728 BARD1; NbExp=9; IntAct=EBI-349905, EBI-473181; P10415 BCL2; NbExp=6; IntAct=EBI-349905, EBI-77694; Q7Z569 BRAP; NbExp=3; IntAct=EBI-349905, EBI-349900; Q6PJG6 BRAT1; NbExp=6; IntAct=EBI-349905, EBI-10826195; Q9BX63 BRIP1; NbExp=12; IntAct=EBI-349905, EBI-3509650; P24385 CCND1; NbExp=3; IntAct=EBI-349905, EBI-375001; P24864 CCNE1; NbExp=2; IntAct=EBI-349905, EBI-519526; O14757 CHEK1; NbExp=3; IntAct=EBI-349905, EBI-974488; P03372 ESR1; NbExp=12; IntAct=EBI-349905, EBI-78473; Q61188 Ezh2 (xeno); NbExp=5; IntAct=EBI-349905, EBI-904311; Q6UWZ7 FAM175A; NbExp=10; IntAct=EBI-349905, EBI-1263451; Q14192 FHL2; NbExp=6; IntAct=EBI-349905, EBI-701903; P78347 GTF2I; NbExp=5; IntAct=EBI-349905, EBI-359622; P16104 H2AFX; NbExp=4; IntAct=EBI-349905, EBI-494830; P10809 HSPD1; NbExp=2; IntAct=EBI-349905, EBI-352528; Q16666 IFI16; NbExp=9; IntAct=EBI-349905, EBI-2867186; P52292 KPNA2; NbExp=3; IntAct=EBI-349905, EBI-349938; Q8WX92 NELFB; NbExp=5; IntAct=EBI-349905, EBI-347721; P62136 PPP1CA; NbExp=2; IntAct=EBI-349905, EBI-357253; P62140 PPP1CB; NbExp=3; IntAct=EBI-349905, EBI-352350; P36873 PPP1CC; NbExp=2; IntAct=EBI-349905, EBI-356283; Q99708 RBBP8; NbExp=9; IntAct=EBI-349905, EBI-745715; Q9Y4A5 TRRAP; NbExp=8; IntAct=EBI-349905, EBI-399128; Q96RL1 UIMC1; NbExp=9; IntAct=EBI-349905, EBI-725300; Q6NZY4 ZCCHC8; NbExp=2; IntAct=EBI-349905, EBI-1263058; Q9GZX5 ZNF350; NbExp=3; IntAct=EBI-349905, EBI-396421; |
| # IntAct | P38398 | 76 |
| # InterPro | IPR001357 | BRCT_dom |
| # InterPro | IPR001841 | Znf_RING |
| # InterPro | IPR011364 | BRCA1 |
| # InterPro | IPR013083 | Znf_RING/FYVE/PHD |
| # InterPro | IPR017907 | Znf_RING_CS |
| # InterPro | IPR018957 | Znf_C3HC4_RING-type |
| # InterPro | IPR025994 | BRCA1_serine_dom |
| # InterPro | IPR031099 | BRCA1-associated |
| # KEGG_Brite | ko00001 | KEGG Orthology (KO) |
| # KEGG_Brite | ko00002 | KEGG pathway modules |
| # KEGG_Brite | ko03036 | Chromosome |
| # KEGG_Brite | ko03400 | DNA repair and recombination proteins |
| # KEGG_Brite | ko03400 | M00295 BRCA1-associated genome surveillance complex (BASC) |
| # KEGG_Brite | ko04121 | Ubiquitin system |
| # KEGG_Disease | H00027 | [Cancer] Ovarian cancer |
| # KEGG_Disease | H00031 | [Cancer] Breast cancer |
| # KEGG_Disease | H01554 | [Cancer] Fallopian tube cancer |
| # KEGG_Disease | H01665 | [Cancer] Primary peritoneal carcinoma |
| # KEGG_Pathway | ko03460 | Fanconi anemia pathway |
| # KEGG_Pathway | ko04120 | Ubiquitin mediated proteolysis |
| # KEGG_Pathway | ko04151 | PI3K-Akt signaling pathway |
| # MIM | 113705 | gene |
| # MIM | 114480 | phenotype |
| # MIM | 167000 | phenotype |
| # MIM | 604370 | phenotype |
| # MIM | 614320 | phenotype |
| # Organism | BRCA1_HUMAN | Homo sapiens (Human) |
| # Orphanet | 1331 | Familial prostate cancer |
| # Orphanet | 1333 | Familial pancreatic carcinoma |
| # Orphanet | 145 | Hereditary breast and ovarian cancer syndrome |
| # Orphanet | 168829 | Primary peritoneal carcinoma |
| # Orphanet | 213524 | Hereditary site-specific ovarian cancer syndrome |
| # Orphanet | 227535 | Hereditary breast cancer |
| # PANTHER | PTHR13763:SF0 | PTHR13763:SF0 |
| # PANTHER | PTHR13763 | PTHR13763 |
| # PATHWAY | BRCA1_HUMAN | Protein modification; protein ubiquitination. |
| # PDB | 1JM7 | NMR; -; A=1-110 |
| # PDB | 1JNX | X-ray; 2.50 A; X=1646-1859 |
| # PDB | 1N5O | X-ray; 2.80 A; X=1646-1859 |
| # PDB | 1OQA | NMR; -; A=1755-1863 |
| # PDB | 1T15 | X-ray; 1.85 A; A=1646-1859 |
| # PDB | 1T29 | X-ray; 2.30 A; A=1646-1859 |
| # PDB | 1T2U | X-ray; 2.80 A; A=1646-1859 |
| # PDB | 1T2V | X-ray; 3.30 A; A/B/C/D/E=1646-1859 |
| # PDB | 1Y98 | X-ray; 2.50 A; A=1646-1859 |
| # PDB | 2ING | X-ray; 3.60 A; X=1649-1859 |
| # PDB | 3COJ | X-ray; 3.21 A; A/B/C/D/E/F/G/X=1646-1859 |
| # PDB | 3K0H | X-ray; 2.70 A; A=1646-1859 |
| # PDB | 3K0K | X-ray; 2.70 A; A=1646-1859 |
| # PDB | 3K15 | X-ray; 2.80 A; A=1646-1859 |
| # PDB | 3K16 | X-ray; 3.00 A; A=1646-1859 |
| # PDB | 3PXA | X-ray; 2.55 A; A=1646-1859 |
| # PDB | 3PXB | X-ray; 2.50 A; A=1646-1859 |
| # PDB | 3PXC | X-ray; 2.80 A; X=1646-1859 |
| # PDB | 3PXD | X-ray; 2.80 A; A=1646-1859 |
| # PDB | 3PXE | X-ray; 2.85 A; A/B/C/D=1646-1859 |
| # PDB | 4IFI | X-ray; 2.20 A; A=1646-1859 |
| # PDB | 4IGK | X-ray; 1.75 A; A/B=1646-1859 |
| # PDB | 4JLU | X-ray; 3.50 A; A=1649-1859 |
| # PDB | 4OFB | X-ray; 3.05 A; A=1646-1859 |
| # PDB | 4U4A | X-ray; 3.51 A; A/B/C=1646-1859 |
| # PDB | 4Y18 | X-ray; 3.50 A; A/B/C/D/E/F/G/H=1646-1859 |
| # PDB | 4Y2G | X-ray; 2.50 A; A=1646-1859 |
| # PIR | A58881 | A58881 |
| # PIRSF | PIRSF001734 | BRCA1 |
| # POLYMORPHISM | BRCA1_HUMAN | There is evidence that the presence of the rare form of Gln-356-Arg and Leu-871-Pro polymorphisms may be associated with an increased risk for developing ovarian cancer. |
| # PRINTS | PR00493 | BRSTCANCERI |
| # PROSITE | PS00518 | ZF_RING_1 |
| # PROSITE | PS50089 | ZF_RING_2 |
| # PROSITE | PS50172 | BRCT; 2 |
| # PTM | BRCA1_HUMAN | Autoubiquitinated, undergoes 'Lys-6'-linked polyubiquitination. 'Lys-6'-linked polyubiquitination does not promote degradation. {ECO 0000269|PubMed 12890688, ECO 0000269|PubMed 20351172}. |
| # PTM | BRCA1_HUMAN | Phosphorylation at Ser-308 by AURKA is required for normal cell cycle progression from G2 to mitosis. Phosphorylated in response to IR, UV, and various stimuli that cause checkpoint activation, probably by ATM or ATR. Phosphorylation at Ser-988 by CHEK2 regulates mitotic spindle assembly. {ECO 0000269|PubMed 10724175, ECO 0000269|PubMed 11114888, ECO 0000269|PubMed 12183412, ECO 0000269|PubMed 14990569, ECO 0000269|PubMed 18056443, ECO 0000269|PubMed 20364141, ECO 0000269|PubMed 21144835}. |
| # Pfam | PF00097 | zf-C3HC4 |
| # Pfam | PF00533 | BRCT; 2 |
| # Pfam | PF12820 | BRCT_assoc |
| # Proteomes | UP000005640 | Chromosome 17 |
| # Reactome | R-HSA-1221632 | Meiotic synapsis |
| # Reactome | R-HSA-3108214 | SUMOylation of DNA damage response and repair proteins |
| # Reactome | R-HSA-5685938 | HDR through Single Strand Annealing (SSA) |
| # Reactome | R-HSA-5685942 | HDR through Homologous Recombination (HRR) |
| # Reactome | R-HSA-5689901 | Metalloprotease DUBs |
| # Reactome | R-HSA-5693554 | Resolution of D-loop Structures through Synthesis-Dependent Strand Annealing (SDSA) |
| # Reactome | R-HSA-5693565 | Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks |
| # Reactome | R-HSA-5693568 | Resolution of D-loop Structures through Holliday Junction Intermediates |
| # Reactome | R-HSA-5693571 | Nonhomologous End-Joining (NHEJ) |
| # Reactome | R-HSA-5693579 | Homologous DNA Pairing and Strand Exchange |
| # Reactome | R-HSA-5693607 | Processing of DNA double-strand break ends |
| # Reactome | R-HSA-5693616 | Presynaptic phase of homologous DNA pairing and strand exchange |
| # Reactome | R-HSA-6796648 | TP53 Regulates Transcription of DNA Repair Genes |
| # Reactome | R-HSA-6804756 | Regulation of TP53 Activity through Phosphorylation |
| # Reactome | R-HSA-69473 | G2/M DNA damage checkpoint |
| # Reactome | R-HSA-912446 | Meiotic recombination |
| # RecName | BRCA1_HUMAN | Breast cancer type 1 susceptibility protein |
| # RefSeq | NP_009225 | NM_007294.3. [P38398-1] |
| # RefSeq | NP_009228 | NM_007297.3. [P38398-8] |
| # RefSeq | NP_009229 | NM_007298.3. [P38398-3] |
| # RefSeq | NP_009230 | NM_007299.3. [P38398-6] |
| # RefSeq | NP_009231 | NM_007300.3. [P38398-7] |
| # SEQUENCE CAUTION | Sequence=AAB61673.1; Type=Erroneous translation; Note=Wrong choice of CDS.; Evidence={ECO:0000305}; Sequence=AAI15038.1; Type=Erroneous initiation; Note=Translation N-terminally extended.; Evidence={ECO | 0000305}; Sequence=AAI15038.1; Type=Erroneous termination; Positions=526; Note=Translated as Gln.; Evidence={ECO:0000305}; |
| # SIMILARITY | Contains 1 RING-type zinc finger. {ECO:0000255|PROSITE-ProRule | PRU00175}. |
| # SIMILARITY | Contains 2 BRCT domains. {ECO:0000255|PROSITE- ProRule | PRU00033}. |
| # SMART | SM00184 | RING |
| # SMART | SM00292 | BRCT; 2 |
| # SUBCELLULAR LOCATION | BRCA1_HUMAN | Isoform 3 Cytoplasm. |
| # SUBCELLULAR LOCATION | BRCA1_HUMAN | Isoform 5 Cytoplasm {ECO 0000269|PubMed 8972225}. |
| # SUBCELLULAR LOCATION | BRCA1_HUMAN | Nucleus {ECO 0000269|PubMed 15133502, ECO 0000269|PubMed 17525340, ECO 0000269|PubMed 20160719, ECO 0000269|PubMed 21144835, ECO 0000269|PubMed 26778126, ECO 0000269|PubMed 9528852}. Chromosome {ECO 0000250|UniProtKB P48754}. Cytoplasm {ECO 0000269|PubMed 20160719}. Note=Localizes at sites of DNA damage at double-strand breaks (DSBs); recruitment to DNA damage sites is mediated by FAM175A and the BRCA1-A complex (PubMed 26778126). Translocated to the cytoplasm during UV-induced apoptosis (PubMed 20160719). {ECO 0000269|PubMed 20160719, ECO 0000269|PubMed 26778126}. |
| # SUBUNIT | BRCA1_HUMAN | Heterodimer with BARD1. Part of the BRCA1-associated genome surveillance complex (BASC), which contains BRCA1, MSH2, MSH6, MLH1, ATM, BLM, PMS2 and the MRE11-RAD50-NBN protein (MRN) complex. This association could be a dynamic process changing throughout the cell cycle and within subnuclear domains. Component of the BRCA1-A complex, at least composed of BRCA1, BARD1, UIMC1/RAP80, FAM175A/Abraxas, BRCC3/BRCC36, BRE/BRCC45 and BABAM1/NBA1. Interacts (via the BRCT domains) with FAM175A (phosphorylated form); this is important for recruitment to sites of DNA damage. Can form a heterotetramer with two molecules of FAM175A (phosphorylated form). Component of the BRCA1-RBBP8 complex. Interacts (via the BRCT domains) with RBBP8 ('Ser-327' phosphorylated form); the interaction ubiquitinates RBBP8, regulates CHEK1 activation, and involves RBBP8 in BRCA1-dependent G2/M checkpoint control on DNA damage. Associates with RNA polymerase II holoenzyme. Interacts with SMC1A, COBRA1, DCLRE1C, CLSPN. CHEK1, CHEK2, BAP1, BRCC3, AURKA, UBXN1 and KIAA0101/PAF15. Interacts (via BRCT domains) with BRIP1 (phosphorylated form). Interacts with FANCD2 (ubiquitinated form). Interacts with H2AFX (phosphorylated on 'Ser-140'). Interacts (via the BRCT domains) with ACACA (phosphorylated form); the interaction prevents dephosphorylation of ACACA. Part of a BRCA complex containing BRCA1, BRCA2 and PALB2. Interacts directly with PALB2; the interaction is essential for its function in HRR. Interacts directly with BRCA2; the interaction occurs only in the presence of PALB2 which serves as the bridging protein. Interacts (via the BRCT domains) with LMO4; the interaction represses the transcriptional activity of BRCA1. Interacts (via the BRCT domains) with CCAR2 (via N-terminus); the interaction represses the transcriptional activator activity of BRCA1. Interacts with EXD2 (PubMed 26807646). {ECO 0000269|PubMed 10724175, ECO 0000269|PubMed 10783165, ECO 0000269|PubMed 11239454, ECO 0000269|PubMed 11301010, ECO 0000269|PubMed 11573085, ECO 0000269|PubMed 11739404, ECO 0000269|PubMed 11751867, ECO 0000269|PubMed 11836499, ECO 0000269|PubMed 11877377, ECO 0000269|PubMed 12360400, ECO 0000269|PubMed 12419185, ECO 0000269|PubMed 12890688, ECO 0000269|PubMed 14636569, ECO 0000269|PubMed 14976165, ECO 0000269|PubMed 14990569, ECO 0000269|PubMed 15096610, ECO 0000269|PubMed 15133502, ECO 0000269|PubMed 15456891, ECO 0000269|PubMed 16101277, ECO 0000269|PubMed 16326698, ECO 0000269|PubMed 16698035, ECO 0000269|PubMed 16818604, ECO 0000269|PubMed 17525340, ECO 0000269|PubMed 17643121, ECO 0000269|PubMed 17643122, ECO 0000269|PubMed 18452305, ECO 0000269|PubMed 19261746, ECO 0000269|PubMed 19261748, ECO 0000269|PubMed 19261749, ECO 0000269|PubMed 19369211, ECO 0000269|PubMed 20159462, ECO 0000269|PubMed 20160719, ECO 0000269|PubMed 20351172, ECO 0000269|PubMed 21473589, ECO 0000269|PubMed 21673012, ECO 0000269|PubMed 24316840, ECO 0000269|PubMed 26778126, ECO 0000269|PubMed 26807646, ECO 0000269|PubMed 9528852, ECO 0000269|PubMed 9811458}. |
| # SUPFAM | SSF52113 | SSF52113; 2 |
| # TISSUE SPECIFICITY | BRCA1_HUMAN | Isoform 1 and isoform 3 are widely expressed. Isoform 3 is reduced or absent in several breast and ovarian cancer cell lines. |
| # UCSC | uc002icq | human. [P38398-1] |
| # UCSC | uc010cyx | human |
| # WEB RESOURCE | BRCA1_HUMAN | Name=Atlas of Genetics and Cytogenetics in Oncology and Haematology; URL="http //atlasgeneticsoncology.org/Genes/BRCA1ID163ch17q21.html"; |
| # WEB RESOURCE | BRCA1_HUMAN | Name=NIEHS-SNPs; URL="http //egp.gs.washington.edu/data/brca1/"; |
| # WEB RESOURCE | BRCA1_HUMAN | Name=SHMPD; Note=The Singapore human mutation and polymorphism database; URL="http //shmpd.bii.a-star.edu.sg/gene.php?genestart=A&genename=BRCA1"; |
| # WEB RESOURCE | BRCA1_HUMAN | Name=Wikipedia; Note=BRCA1 entry; URL="https //en.wikipedia.org/wiki/BRCA1"; |
| # eggNOG | ENOG410ITQ4 | Eukaryota |
| # eggNOG | ENOG4112BIH | LUCA |
| BLAST | swissprot:BRCA1_HUMAN | http://rest.g-language.org/emboss/kblast/swissprot:BRCA1_HUMAN |
| BioCyc | ZFISH:ENSG00000012048-MONOMER | http://biocyc.org/getid?id=ZFISH:ENSG00000012048-MONOMER |
| COXPRESdb | 672 | http://coxpresdb.jp/data/gene/672.shtml |
| CleanEx | HS_BRCA1 | http://www.cleanex.isb-sib.ch/cgi-bin/get_doc?db=cleanex&format=nice&entry=HS_BRCA1 |
| DIP | DIP-5971N | http://dip.doe-mbi.ucla.edu/dip/Browse.cgi?ID=DIP-5971N |
| DOI | 10.1002/(SICI)1098-1004(1996)7:4<334::AID-HUMU7>3.3.CO | 3.3.CO>http://dx.doi.org/10.1002/(SICI)1098-1004(1996)7:4<334::AID-HUMU7>3.3.CO |
| DOI | 10.1002/(SICI)1098-1004(1998)11:2<166::AID-HUMU10>3.0.CO | 3.0.CO>http://dx.doi.org/10.1002/(SICI)1098-1004(1998)11:2<166::AID-HUMU10>3.0.CO |
| DOI | 10.1002/(SICI)1098-2272(1996)13:6<595::AID-GEPI5>3.3.CO | 3.3.CO>http://dx.doi.org/10.1002/(SICI)1098-2272(1996)13:6<595::AID-GEPI5>3.3.CO |
| DOI | 10.1002/humu.1380080102 | http://dx.doi.org/10.1002/humu.1380080102 |
| DOI | 10.1002/humu.21267 | http://dx.doi.org/10.1002/humu.21267 |
| DOI | 10.1002/humu.9083 | http://dx.doi.org/10.1002/humu.9083 |
| DOI | 10.1002/humu.9084 | http://dx.doi.org/10.1002/humu.9084 |
| DOI | 10.1002/humu.9174 | http://dx.doi.org/10.1002/humu.9174 |
| DOI | 10.1002/humu.9213 | http://dx.doi.org/10.1002/humu.9213 |
| DOI | 10.1002/humu.9275 | http://dx.doi.org/10.1002/humu.9275 |
| DOI | 10.1007/s00439-008-0554-0 | http://dx.doi.org/10.1007/s00439-008-0554-0 |
| DOI | 10.1007/s004390050799 | http://dx.doi.org/10.1007/s004390050799 |
| DOI | 10.1007/s004390050936 | http://dx.doi.org/10.1007/s004390050936 |
| DOI | 10.1007/s100380050035 | http://dx.doi.org/10.1007/s100380050035 |
| DOI | 10.1016/S0092-8674(01)00304-X | http://dx.doi.org/10.1016/S0092-8674(01)00304-X |
| DOI | 10.1016/S0960-9822(02)01259-9 | http://dx.doi.org/10.1016/S0960-9822(02)01259-9 |
| DOI | 10.1016/S1097-2765(01)00173-3 | http://dx.doi.org/10.1016/S1097-2765(01)00173-3 |
| DOI | 10.1016/S1097-2765(03)00281-8 | http://dx.doi.org/10.1016/S1097-2765(03)00281-8 |
| DOI | 10.1016/S1097-2765(03)00424-6 | http://dx.doi.org/10.1016/S1097-2765(03)00424-6 |
| DOI | 10.1016/j.bbrc.2010.12.005 | http://dx.doi.org/10.1016/j.bbrc.2010.12.005 |
| DOI | 10.1016/j.cell.2006.09.026 | http://dx.doi.org/10.1016/j.cell.2006.09.026 |
| DOI | 10.1016/j.ejca.2003.09.016 | http://dx.doi.org/10.1016/j.ejca.2003.09.016 |
| DOI | 10.1016/j.jmb.2006.04.010 | http://dx.doi.org/10.1016/j.jmb.2006.04.010 |
| DOI | 10.1016/j.molcel.2015.12.017 | http://dx.doi.org/10.1016/j.molcel.2015.12.017 |
| DOI | 10.1016/j.str.2009.12.008 | http://dx.doi.org/10.1016/j.str.2009.12.008 |
| DOI | 10.1021/bi049550q | http://dx.doi.org/10.1021/bi049550q |
| DOI | 10.1021/bi0509651 | http://dx.doi.org/10.1021/bi0509651 |
| DOI | 10.1021/bi2003795 | http://dx.doi.org/10.1021/bi2003795 |
| DOI | 10.1021/bi800314m | http://dx.doi.org/10.1021/bi800314m |
| DOI | 10.1021/pr300630k | http://dx.doi.org/10.1021/pr300630k |
| DOI | 10.1038/35004614 | http://dx.doi.org/10.1038/35004614 |
| DOI | 10.1038/ejhg.2008.13 | http://dx.doi.org/10.1038/ejhg.2008.13 |
| DOI | 10.1038/nature04689 | http://dx.doi.org/10.1038/nature04689 |
| DOI | 10.1038/ncb2051 | http://dx.doi.org/10.1038/ncb2051 |
| DOI | 10.1038/ncb3303 | http://dx.doi.org/10.1038/ncb3303 |
| DOI | 10.1038/ng1294-387 | http://dx.doi.org/10.1038/ng1294-387 |
| DOI | 10.1038/ng1294-399 | http://dx.doi.org/10.1038/ng1294-399 |
| DOI | 10.1038/ng837 | http://dx.doi.org/10.1038/ng837 |
| DOI | 10.1038/nsb1001-833 | http://dx.doi.org/10.1038/nsb1001-833 |
| DOI | 10.1038/nsb1001-838 | http://dx.doi.org/10.1038/nsb1001-838 |
| DOI | 10.1038/nsmb.2890 | http://dx.doi.org/10.1038/nsmb.2890 |
| DOI | 10.1038/nsmb1277 | http://dx.doi.org/10.1038/nsmb1277 |
| DOI | 10.1038/nsmb1279 | http://dx.doi.org/10.1038/nsmb1279 |
| DOI | 10.1038/nsmb775 | http://dx.doi.org/10.1038/nsmb775 |
| DOI | 10.1038/sj.bjc.6601656 | http://dx.doi.org/10.1038/sj.bjc.6601656 |
| DOI | 10.1038/sj.bjc.6605577 | http://dx.doi.org/10.1038/sj.bjc.6605577 |
| DOI | 10.1038/sj.onc.1200924 | http://dx.doi.org/10.1038/sj.onc.1200924 |
| DOI | 10.1038/sj.onc.1201861 | http://dx.doi.org/10.1038/sj.onc.1201861 |
| DOI | 10.1038/sj.onc.1202150 | http://dx.doi.org/10.1038/sj.onc.1202150 |
| DOI | 10.1038/sj.onc.1203599 | http://dx.doi.org/10.1038/sj.onc.1203599 |
| DOI | 10.1038/sj.onc.1205915 | http://dx.doi.org/10.1038/sj.onc.1205915 |
| DOI | 10.1073/pnas.0401847101 | http://dx.doi.org/10.1073/pnas.0401847101 |
| DOI | 10.1073/pnas.0805139105 | http://dx.doi.org/10.1073/pnas.0805139105 |
| DOI | 10.1073/pnas.0811159106 | http://dx.doi.org/10.1073/pnas.0811159106 |
| DOI | 10.1073/pnas.1210303109 | http://dx.doi.org/10.1073/pnas.1210303109 |
| DOI | 10.1073/pnas.96.20.11364 | http://dx.doi.org/10.1073/pnas.96.20.11364 |
| DOI | 10.1074/jbc.C300249200 | http://dx.doi.org/10.1074/jbc.C300249200 |
| DOI | 10.1074/jbc.M110603200 | http://dx.doi.org/10.1074/jbc.M110603200 |
| DOI | 10.1074/jbc.M210019200 | http://dx.doi.org/10.1074/jbc.M210019200 |
| DOI | 10.1074/jbc.M311780200 | http://dx.doi.org/10.1074/jbc.M311780200 |
| DOI | 10.1074/jbc.M504652200 | http://dx.doi.org/10.1074/jbc.M504652200 |
| DOI | 10.1074/mcp.O114.044792 | http://dx.doi.org/10.1074/mcp.O114.044792 |
| DOI | 10.1083/jcb.200108049 | http://dx.doi.org/10.1083/jcb.200108049 |
| DOI | 10.1086/302583 | http://dx.doi.org/10.1086/302583 |
| DOI | 10.1086/521032 | http://dx.doi.org/10.1086/521032 |
| DOI | 10.1089/10906570260199375 | http://dx.doi.org/10.1089/10906570260199375 |
| DOI | 10.1093/hmg/5.6.835 | http://dx.doi.org/10.1093/hmg/5.6.835 |
| DOI | 10.1093/hmg/8.5.889 | http://dx.doi.org/10.1093/hmg/8.5.889 |
| DOI | 10.1093/hmg/ddh095 | http://dx.doi.org/10.1093/hmg/ddh095 |
| DOI | 10.1101/gad.1431006 | http://dx.doi.org/10.1101/gad.1431006 |
| DOI | 10.1101/gad.1739609 | http://dx.doi.org/10.1101/gad.1739609 |
| DOI | 10.1101/gad.1770309 | http://dx.doi.org/10.1101/gad.1770309 |
| DOI | 10.1101/gad.1770609 | http://dx.doi.org/10.1101/gad.1770609 |
| DOI | 10.1101/gad.851000 | http://dx.doi.org/10.1101/gad.851000 |
| DOI | 10.1101/gad.970702 | http://dx.doi.org/10.1101/gad.970702 |
| DOI | 10.1101/gr.2596504 | http://dx.doi.org/10.1101/gr.2596504 |
| DOI | 10.1101/gr.6.11.1029 | http://dx.doi.org/10.1101/gr.6.11.1029 |
| DOI | 10.1107/S1744309113030649 | http://dx.doi.org/10.1107/S1744309113030649 |
| DOI | 10.1126/science.1133427 | http://dx.doi.org/10.1126/science.1133427 |
| DOI | 10.1126/science.1139476 | http://dx.doi.org/10.1126/science.1139476 |
| DOI | 10.1126/science.1140321 | http://dx.doi.org/10.1126/science.1140321 |
| DOI | 10.1126/science.7545954 | http://dx.doi.org/10.1126/science.7545954 |
| DOI | 10.1126/science.7939630 | http://dx.doi.org/10.1126/science.7939630 |
| DOI | 10.1126/scisignal.2000007 | http://dx.doi.org/10.1126/scisignal.2000007 |
| DOI | 10.1126/scisignal.2000475 | http://dx.doi.org/10.1126/scisignal.2000475 |
| DOI | 10.1126/scisignal.2001570 | http://dx.doi.org/10.1126/scisignal.2001570 |
| DOI | 10.1128/MCB.01056-09 | http://dx.doi.org/10.1128/MCB.01056-09 |
| DOI | 10.1128/MCB.17.1.444 | http://dx.doi.org/10.1128/MCB.17.1.444 |
| DOI | 10.1128/MCB.24.20.9207-9220.2004 | http://dx.doi.org/10.1128/MCB.24.20.9207-9220.2004 |
| DOI | 10.1158/0008-5472.CAN-07-2578 | http://dx.doi.org/10.1158/0008-5472.CAN-07-2578 |
| DOI | 10.1158/1541-7786.MCR-10-0503 | http://dx.doi.org/10.1158/1541-7786.MCR-10-0503 |
| DOI | 10.1158/2159-8290.CD-13-0094 | http://dx.doi.org/10.1158/2159-8290.CD-13-0094 |
| DisProt | DP00238 | http://www.disprot.org/protein.php?id=DP00238 |
| EC_number | EC:2.3.2.27 {ECO:0000269|PubMed:10500182, ECO:0000269|PubMed:12887909, ECO:0000269|PubMed:12890688, ECO:0000269|PubMed:16818604, ECO:0000269|PubMed:18056443, ECO:0000269|PubMed:20351172} | http://www.genome.jp/dbget-bin/www_bget?EC:2.3.2.27 {ECO:0000269|PubMed:10500182, ECO:0000269|PubMed:12887909, ECO:0000269|PubMed:12890688, ECO:0000269|PubMed:16818604, ECO:0000269|PubMed:18056443, ECO:0000269|PubMed:20351172} |
| EMBL | AC060780 | http://www.ebi.ac.uk/ena/data/view/AC060780 |
| EMBL | AC135721 | http://www.ebi.ac.uk/ena/data/view/AC135721 |
| EMBL | AF005068 | http://www.ebi.ac.uk/ena/data/view/AF005068 |
| EMBL | AY273801 | http://www.ebi.ac.uk/ena/data/view/AY273801 |
| EMBL | BC072418 | http://www.ebi.ac.uk/ena/data/view/BC072418 |
| EMBL | BC115037 | http://www.ebi.ac.uk/ena/data/view/BC115037 |
| EMBL | DQ190450 | http://www.ebi.ac.uk/ena/data/view/DQ190450 |
| EMBL | DQ190451 | http://www.ebi.ac.uk/ena/data/view/DQ190451 |
| EMBL | DQ190452 | http://www.ebi.ac.uk/ena/data/view/DQ190452 |
| EMBL | DQ190453 | http://www.ebi.ac.uk/ena/data/view/DQ190453 |
| EMBL | DQ190454 | http://www.ebi.ac.uk/ena/data/view/DQ190454 |
| EMBL | DQ190455 | http://www.ebi.ac.uk/ena/data/view/DQ190455 |
| EMBL | DQ190456 | http://www.ebi.ac.uk/ena/data/view/DQ190456 |
| EMBL | L78833 | http://www.ebi.ac.uk/ena/data/view/L78833 |
| EMBL | U14680 | http://www.ebi.ac.uk/ena/data/view/U14680 |
| EMBL | U64805 | http://www.ebi.ac.uk/ena/data/view/U64805 |
| ENZYME | 2.3.2.27 {ECO:0000269|PubMed:10500182, ECO:0000269|PubMed:12887909, ECO:0000269|PubMed:12890688, ECO:0000269|PubMed:16818604, ECO:0000269|PubMed:18056443, ECO:0000269|PubMed:20351172} | http://enzyme.expasy.org/EC/2.3.2.27 {ECO:0000269|PubMed:10500182, ECO:0000269|PubMed:12887909, ECO:0000269|PubMed:12890688, ECO:0000269|PubMed:16818604, ECO:0000269|PubMed:18056443, ECO:0000269|PubMed:20351172} |
| Ensembl | ENST00000352993 | http://www.ensembl.org/id/ENST00000352993 |
| Ensembl | ENST00000357654 | http://www.ensembl.org/id/ENST00000357654 |
| Ensembl | ENST00000461221 | http://www.ensembl.org/id/ENST00000461221 |
| Ensembl | ENST00000461798 | http://www.ensembl.org/id/ENST00000461798 |
| Ensembl | ENST00000468300 | http://www.ensembl.org/id/ENST00000468300 |
| Ensembl | ENST00000471181 | http://www.ensembl.org/id/ENST00000471181 |
| Ensembl | ENST00000491747 | http://www.ensembl.org/id/ENST00000491747 |
| Ensembl | ENST00000493795 | http://www.ensembl.org/id/ENST00000493795 |
| G-Links | BRCA1_HUMAN | http://link.g-language.org/BRCA1_HUMAN/format=tsv |
| GO_component | GO:0000151 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0000151 |
| GO_component | GO:0000800 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0000800 |
| GO_component | GO:0005634 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0005634 |
| GO_component | GO:0005654 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0005654 |
| GO_component | GO:0005694 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0005694 |
| GO_component | GO:0005737 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0005737 |
| GO_component | GO:0005886 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0005886 |
| GO_component | GO:0008274 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0008274 |
| GO_component | GO:0030529 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0030529 |
| GO_component | GO:0031436 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0031436 |
| GO_component | GO:0043234 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0043234 |
| GO_component | GO:0070531 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0070531 |
| GO_function | GO:0003677 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0003677 |
| GO_function | GO:0003684 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0003684 |
| GO_function | GO:0003713 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0003713 |
| GO_function | GO:0003723 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0003723 |
| GO_function | GO:0004842 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0004842 |
| GO_function | GO:0008270 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0008270 |
| GO_function | GO:0015631 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0015631 |
| GO_function | GO:0019899 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0019899 |
| GO_function | GO:0031625 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0031625 |
| GO_function | GO:0044212 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0044212 |
| GO_function | GO:0050681 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0050681 |
| GO_process | GO:0000724 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0000724 |
| GO_process | GO:0000729 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0000729 |
| GO_process | GO:0000731 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0000731 |
| GO_process | GO:0000732 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0000732 |
| GO_process | GO:0006260 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0006260 |
| GO_process | GO:0006301 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0006301 |
| GO_process | GO:0006302 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0006302 |
| GO_process | GO:0006303 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0006303 |
| GO_process | GO:0006349 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0006349 |
| GO_process | GO:0006351 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0006351 |
| GO_process | GO:0006357 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0006357 |
| GO_process | GO:0006359 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0006359 |
| GO_process | GO:0006633 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0006633 |
| GO_process | GO:0006915 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0006915 |
| GO_process | GO:0006974 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0006974 |
| GO_process | GO:0006978 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0006978 |
| GO_process | GO:0007059 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0007059 |
| GO_process | GO:0007098 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0007098 |
| GO_process | GO:0008630 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0008630 |
| GO_process | GO:0009048 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0009048 |
| GO_process | GO:0010212 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0010212 |
| GO_process | GO:0010575 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0010575 |
| GO_process | GO:0010628 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0010628 |
| GO_process | GO:0016567 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0016567 |
| GO_process | GO:0016925 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0016925 |
| GO_process | GO:0030521 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0030521 |
| GO_process | GO:0031052 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0031052 |
| GO_process | GO:0031398 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0031398 |
| GO_process | GO:0031572 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0031572 |
| GO_process | GO:0033147 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0033147 |
| GO_process | GO:0035066 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0035066 |
| GO_process | GO:0035067 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0035067 |
| GO_process | GO:0042127 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0042127 |
| GO_process | GO:0042981 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0042981 |
| GO_process | GO:0043009 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0043009 |
| GO_process | GO:0043627 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0043627 |
| GO_process | GO:0044030 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0044030 |
| GO_process | GO:0045717 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0045717 |
| GO_process | GO:0045739 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0045739 |
| GO_process | GO:0045766 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0045766 |
| GO_process | GO:0045892 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0045892 |
| GO_process | GO:0045893 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0045893 |
| GO_process | GO:0045944 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0045944 |
| GO_process | GO:0046600 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0046600 |
| GO_process | GO:0051571 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0051571 |
| GO_process | GO:0051572 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0051572 |
| GO_process | GO:0051573 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0051573 |
| GO_process | GO:0051574 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0051574 |
| GO_process | GO:0051865 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0051865 |
| GO_process | GO:0070512 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0070512 |
| GO_process | GO:0071158 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0071158 |
| GO_process | GO:0071356 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0071356 |
| GO_process | GO:0071681 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0071681 |
| GO_process | GO:0085020 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0085020 |
| GO_process | GO:1901796 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:1901796 |
| GO_process | GO:1902042 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:1902042 |
| GO_process | GO:2000378 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:2000378 |
| GO_process | GO:2000617 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:2000617 |
| GO_process | GO:2000620 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:2000620 |
| GOslim_component | GO:0005575 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0005575 |
| GOslim_component | GO:0005634 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0005634 |
| GOslim_component | GO:0005654 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0005654 |
| GOslim_component | GO:0005694 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0005694 |
| GOslim_component | GO:0005737 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0005737 |
| GOslim_component | GO:0005886 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0005886 |
| GOslim_component | GO:0043234 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0043234 |
| GOslim_function | GO:0000988 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0000988 |
| GOslim_function | GO:0003674 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0003674 |
| GOslim_function | GO:0003677 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0003677 |
| GOslim_function | GO:0003723 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0003723 |
| GOslim_function | GO:0008092 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0008092 |
| GOslim_function | GO:0019899 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0019899 |
| GOslim_function | GO:0043167 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0043167 |
| GOslim_process | GO:0006259 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0006259 |
| GOslim_process | GO:0006464 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0006464 |
| GOslim_process | GO:0006629 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0006629 |
| GOslim_process | GO:0006950 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0006950 |
| GOslim_process | GO:0007059 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0007059 |
| GOslim_process | GO:0007165 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0007165 |
| GOslim_process | GO:0008150 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0008150 |
| GOslim_process | GO:0008219 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0008219 |
| GOslim_process | GO:0009058 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0009058 |
| GOslim_process | GO:0009790 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0009790 |
| GOslim_process | GO:0034641 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0034641 |
| GOslim_process | GO:0044281 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0044281 |
| GOslim_process | GO:0051276 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0051276 |
| Gene3D | 3.30.40.10 | http://www.cathdb.info/version/latest/superfamily/3.30.40.10 |
| Gene3D | 3.40.50.10190 | http://www.cathdb.info/version/latest/superfamily/3.40.50.10190 |
| GeneCards | BRCA1 | http://www.genecards.org/cgi-bin/carddisp.pl?gc_id=BRCA1 |
| GeneID | 672 | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&term=672 |
| GeneTree | ENSGT00440000034289 | http://asia.ensembl.org/Multi/GeneTree/Image?gt=ENSGT00440000034289 |
| HGNC | HGNC:1100 | http://www.genenames.org/data/hgnc_data.php?hgnc_id=HGNC:1100 |
| HOGENOM | HOG000230969 | http://pbil.univ-lyon1.fr/cgi-bin/view-tree.pl?query=HOG000230969&db=HOGENOM6 |
| HOVERGEN | HBG050730 | http://pbil.univ-lyon1.fr/cgi-bin/acnuc-ac2tree?query=HBG050730&db=HOVERGEN |
| HPA | CAB001946 | http://www.proteinatlas.org/tissue_profile.php?antibody_id=CAB001946 |
| HPA | CAB018369 | http://www.proteinatlas.org/tissue_profile.php?antibody_id=CAB018369 |
| HPA | HPA034966 | http://www.proteinatlas.org/tissue_profile.php?antibody_id=HPA034966 |
| InParanoid | P38398 | http://inparanoid.sbc.su.se/cgi-bin/gene_search.cgi?id=P38398 |
| IntAct | P38398 | http://www.ebi.ac.uk/intact/pages/interactions/interactions.xhtml?query=P38398* |
| IntEnz | 2.3.2.27 {ECO:0000269|PubMed:10500182, ECO:0000269|PubMed:12887909, ECO:0000269|PubMed:12890688, ECO:0000269|PubMed:16818604, ECO:0000269|PubMed:18056443, ECO:0000269|PubMed:20351172} | http://www.ebi.ac.uk/intenz/query?cmd=Search&q=2.3.2.27 {ECO:0000269|PubMed:10500182, ECO:0000269|PubMed:12887909, ECO:0000269|PubMed:12890688, ECO:0000269|PubMed:16818604, ECO:0000269|PubMed:18056443, ECO:0000269|PubMed:20351172} |
| InterPro | IPR001357 | http://www.ebi.ac.uk/interpro/entry/IPR001357 |
| InterPro | IPR001841 | http://www.ebi.ac.uk/interpro/entry/IPR001841 |
| InterPro | IPR011364 | http://www.ebi.ac.uk/interpro/entry/IPR011364 |
| InterPro | IPR013083 | http://www.ebi.ac.uk/interpro/entry/IPR013083 |
| InterPro | IPR017907 | http://www.ebi.ac.uk/interpro/entry/IPR017907 |
| InterPro | IPR018957 | http://www.ebi.ac.uk/interpro/entry/IPR018957 |
| InterPro | IPR025994 | http://www.ebi.ac.uk/interpro/entry/IPR025994 |
| InterPro | IPR031099 | http://www.ebi.ac.uk/interpro/entry/IPR031099 |
| Jabion | 672 | http://www.bioportal.jp/genome/cgi-bin/gene_homolog.cgi?org=hs&id=672 |
| KEGG_Brite | ko00001 | http://www.genome.jp/dbget-bin/www_bget?ko00001 |
| KEGG_Brite | ko00002 | http://www.genome.jp/dbget-bin/www_bget?ko00002 |
| KEGG_Brite | ko03036 | http://www.genome.jp/dbget-bin/www_bget?ko03036 |
| KEGG_Brite | ko03400 | http://www.genome.jp/dbget-bin/www_bget?ko03400 |
| KEGG_Brite | ko03400 | http://www.genome.jp/dbget-bin/www_bget?ko03400 |
| KEGG_Brite | ko04121 | http://www.genome.jp/dbget-bin/www_bget?ko04121 |
| KEGG_Disease | H00027 | http://www.genome.jp/dbget-bin/www_bget?H00027 |
| KEGG_Disease | H00031 | http://www.genome.jp/dbget-bin/www_bget?H00031 |
| KEGG_Disease | H01554 | http://www.genome.jp/dbget-bin/www_bget?H01554 |
| KEGG_Disease | H01665 | http://www.genome.jp/dbget-bin/www_bget?H01665 |
| KEGG_Gene | hsa:672 | http://www.genome.jp/dbget-bin/www_bget?hsa:672 |
| KEGG_Orthology | KO:K10605 | http://www.genome.jp/dbget-bin/www_bget?KO:K10605 |
| KEGG_Pathway | ko03460 | http://www.genome.jp/kegg-bin/show_pathway?ko03460 |
| KEGG_Pathway | ko04120 | http://www.genome.jp/kegg-bin/show_pathway?ko04120 |
| KEGG_Pathway | ko04151 | http://www.genome.jp/kegg-bin/show_pathway?ko04151 |
| MIM | 113705 | http://www.ncbi.nlm.nih.gov/omim/113705 |
| MIM | 114480 | http://www.ncbi.nlm.nih.gov/omim/114480 |
| MIM | 167000 | http://www.ncbi.nlm.nih.gov/omim/167000 |
| MIM | 604370 | http://www.ncbi.nlm.nih.gov/omim/604370 |
| MIM | 614320 | http://www.ncbi.nlm.nih.gov/omim/614320 |
| MINT | MINT-90433 | http://mint.bio.uniroma2.it/mint/search/search.do?queryType=protein&interactorAc=MINT-90433 |
| OMA | FQHLLFG | http://omabrowser.org/cgi-bin/gateway.pl?f=DisplayGroup&p1=FQHLLFG |
| Orphanet | 1331 | http://www.orpha.net/consor/cgi-bin/OC_Exp.php?Lng=EN&Expert=1331 |
| Orphanet | 1333 | http://www.orpha.net/consor/cgi-bin/OC_Exp.php?Lng=EN&Expert=1333 |
| Orphanet | 145 | http://www.orpha.net/consor/cgi-bin/OC_Exp.php?Lng=EN&Expert=145 |
| Orphanet | 168829 | http://www.orpha.net/consor/cgi-bin/OC_Exp.php?Lng=EN&Expert=168829 |
| Orphanet | 213524 | http://www.orpha.net/consor/cgi-bin/OC_Exp.php?Lng=EN&Expert=213524 |
| Orphanet | 227535 | http://www.orpha.net/consor/cgi-bin/OC_Exp.php?Lng=EN&Expert=227535 |
| OrthoDB | EOG091G0670 | http://cegg.unige.ch/orthodb/results?searchtext=EOG091G0670 |
| PANTHER | PTHR13763:SF0 | http://www.pantherdb.org/panther/family.do?clsAccession=PTHR13763:SF0 |
| PANTHER | PTHR13763 | http://www.pantherdb.org/panther/family.do?clsAccession=PTHR13763 |
| PDB | 1JM7 | http://www.ebi.ac.uk/pdbe-srv/view/entry/1JM7 |
| PDB | 1JNX | http://www.ebi.ac.uk/pdbe-srv/view/entry/1JNX |
| PDB | 1N5O | http://www.ebi.ac.uk/pdbe-srv/view/entry/1N5O |
| PDB | 1OQA | http://www.ebi.ac.uk/pdbe-srv/view/entry/1OQA |
| PDB | 1T15 | http://www.ebi.ac.uk/pdbe-srv/view/entry/1T15 |
| PDB | 1T29 | http://www.ebi.ac.uk/pdbe-srv/view/entry/1T29 |
| PDB | 1T2U | http://www.ebi.ac.uk/pdbe-srv/view/entry/1T2U |
| PDB | 1T2V | http://www.ebi.ac.uk/pdbe-srv/view/entry/1T2V |
| PDB | 1Y98 | http://www.ebi.ac.uk/pdbe-srv/view/entry/1Y98 |
| PDB | 2ING | http://www.ebi.ac.uk/pdbe-srv/view/entry/2ING |
| PDB | 3COJ | http://www.ebi.ac.uk/pdbe-srv/view/entry/3COJ |
| PDB | 3K0H | http://www.ebi.ac.uk/pdbe-srv/view/entry/3K0H |
| PDB | 3K0K | http://www.ebi.ac.uk/pdbe-srv/view/entry/3K0K |
| PDB | 3K15 | http://www.ebi.ac.uk/pdbe-srv/view/entry/3K15 |
| PDB | 3K16 | http://www.ebi.ac.uk/pdbe-srv/view/entry/3K16 |
| PDB | 3PXA | http://www.ebi.ac.uk/pdbe-srv/view/entry/3PXA |
| PDB | 3PXB | http://www.ebi.ac.uk/pdbe-srv/view/entry/3PXB |
| PDB | 3PXC | http://www.ebi.ac.uk/pdbe-srv/view/entry/3PXC |
| PDB | 3PXD | http://www.ebi.ac.uk/pdbe-srv/view/entry/3PXD |
| PDB | 3PXE | http://www.ebi.ac.uk/pdbe-srv/view/entry/3PXE |
| PDB | 4IFI | http://www.ebi.ac.uk/pdbe-srv/view/entry/4IFI |
| PDB | 4IGK | http://www.ebi.ac.uk/pdbe-srv/view/entry/4IGK |
| PDB | 4JLU | http://www.ebi.ac.uk/pdbe-srv/view/entry/4JLU |
| PDB | 4OFB | http://www.ebi.ac.uk/pdbe-srv/view/entry/4OFB |
| PDB | 4U4A | http://www.ebi.ac.uk/pdbe-srv/view/entry/4U4A |
| PDB | 4Y18 | http://www.ebi.ac.uk/pdbe-srv/view/entry/4Y18 |
| PDB | 4Y2G | http://www.ebi.ac.uk/pdbe-srv/view/entry/4Y2G |
| PDBsum | 1JM7 | http://www.ebi.ac.uk/pdbsum/1JM7 |
| PDBsum | 1JNX | http://www.ebi.ac.uk/pdbsum/1JNX |
| PDBsum | 1N5O | http://www.ebi.ac.uk/pdbsum/1N5O |
| PDBsum | 1OQA | http://www.ebi.ac.uk/pdbsum/1OQA |
| PDBsum | 1T15 | http://www.ebi.ac.uk/pdbsum/1T15 |
| PDBsum | 1T29 | http://www.ebi.ac.uk/pdbsum/1T29 |
| PDBsum | 1T2U | http://www.ebi.ac.uk/pdbsum/1T2U |
| PDBsum | 1T2V | http://www.ebi.ac.uk/pdbsum/1T2V |
| PDBsum | 1Y98 | http://www.ebi.ac.uk/pdbsum/1Y98 |
| PDBsum | 2ING | http://www.ebi.ac.uk/pdbsum/2ING |
| PDBsum | 3COJ | http://www.ebi.ac.uk/pdbsum/3COJ |
| PDBsum | 3K0H | http://www.ebi.ac.uk/pdbsum/3K0H |
| PDBsum | 3K0K | http://www.ebi.ac.uk/pdbsum/3K0K |
| PDBsum | 3K15 | http://www.ebi.ac.uk/pdbsum/3K15 |
| PDBsum | 3K16 | http://www.ebi.ac.uk/pdbsum/3K16 |
| PDBsum | 3PXA | http://www.ebi.ac.uk/pdbsum/3PXA |
| PDBsum | 3PXB | http://www.ebi.ac.uk/pdbsum/3PXB |
| PDBsum | 3PXC | http://www.ebi.ac.uk/pdbsum/3PXC |
| PDBsum | 3PXD | http://www.ebi.ac.uk/pdbsum/3PXD |
| PDBsum | 3PXE | http://www.ebi.ac.uk/pdbsum/3PXE |
| PDBsum | 4IFI | http://www.ebi.ac.uk/pdbsum/4IFI |
| PDBsum | 4IGK | http://www.ebi.ac.uk/pdbsum/4IGK |
| PDBsum | 4JLU | http://www.ebi.ac.uk/pdbsum/4JLU |
| PDBsum | 4OFB | http://www.ebi.ac.uk/pdbsum/4OFB |
| PDBsum | 4U4A | http://www.ebi.ac.uk/pdbsum/4U4A |
| PDBsum | 4Y18 | http://www.ebi.ac.uk/pdbsum/4Y18 |
| PDBsum | 4Y2G | http://www.ebi.ac.uk/pdbsum/4Y2G |
| PRINTS | PR00493 | http://umber.sbs.man.ac.uk/cgi-bin/dbbrowser/sprint/searchprintss.cgi?display_opts=Prints&category=None&queryform=false&prints_accn=PR00493 |
| PROSITE | PS00518 | http://prosite.expasy.org/cgi-bin/prosite/nicedoc.pl?PS00518 |
| PROSITE | PS50089 | http://prosite.expasy.org/cgi-bin/prosite/nicedoc.pl?PS50089 |
| PROSITE | PS50172 | http://prosite.expasy.org/cgi-bin/prosite/nicedoc.pl?PS50172 |
| PSORT-B | swissprot:BRCA1_HUMAN | http://rest.g-language.org/emboss/kpsortb/swissprot:BRCA1_HUMAN |
| PSORT2 | swissprot:BRCA1_HUMAN | http://rest.g-language.org/emboss/kpsort2/swissprot:BRCA1_HUMAN |
| PSORT | swissprot:BRCA1_HUMAN | http://rest.g-language.org/emboss/kpsort/swissprot:BRCA1_HUMAN |
| Pfam | PF00097 | http://pfam.xfam.org/family/PF00097 |
| Pfam | PF00533 | http://pfam.xfam.org/family/PF00533 |
| Pfam | PF12820 | http://pfam.xfam.org/family/PF12820 |
| PharmGKB | PA25411 | http://www.pharmgkb.org/do/serve?objId=PA25411&objCls=Gene |
| Phobius | swissprot:BRCA1_HUMAN | http://rest.g-language.org/emboss/kphobius/swissprot:BRCA1_HUMAN |
| PhylomeDB | P38398 | http://phylomedb.org/?seqid=P38398 |
| ProteinModelPortal | P38398 | http://www.proteinmodelportal.org/query/uniprot/P38398 |
| PubMed | 10196379 | http://www.ncbi.nlm.nih.gov/pubmed/10196379 |
| PubMed | 10323242 | http://www.ncbi.nlm.nih.gov/pubmed/10323242 |
| PubMed | 10486320 | http://www.ncbi.nlm.nih.gov/pubmed/10486320 |
| PubMed | 10500182 | http://www.ncbi.nlm.nih.gov/pubmed/10500182 |
| PubMed | 10724175 | http://www.ncbi.nlm.nih.gov/pubmed/10724175 |
| PubMed | 10783165 | http://www.ncbi.nlm.nih.gov/pubmed/10783165 |
| PubMed | 10851077 | http://www.ncbi.nlm.nih.gov/pubmed/10851077 |
| PubMed | 11114888 | http://www.ncbi.nlm.nih.gov/pubmed/11114888 |
| PubMed | 11239454 | http://www.ncbi.nlm.nih.gov/pubmed/11239454 |
| PubMed | 11301010 | http://www.ncbi.nlm.nih.gov/pubmed/11301010 |
| PubMed | 11573085 | http://www.ncbi.nlm.nih.gov/pubmed/11573085 |
| PubMed | 11573086 | http://www.ncbi.nlm.nih.gov/pubmed/11573086 |
| PubMed | 11739404 | http://www.ncbi.nlm.nih.gov/pubmed/11739404 |
| PubMed | 11751867 | http://www.ncbi.nlm.nih.gov/pubmed/11751867 |
| PubMed | 11836499 | http://www.ncbi.nlm.nih.gov/pubmed/11836499 |
| PubMed | 11877377 | http://www.ncbi.nlm.nih.gov/pubmed/11877377 |
| PubMed | 12183412 | http://www.ncbi.nlm.nih.gov/pubmed/12183412 |
| PubMed | 12215251 | http://www.ncbi.nlm.nih.gov/pubmed/12215251 |
| PubMed | 12360400 | http://www.ncbi.nlm.nih.gov/pubmed/12360400 |
| PubMed | 12419185 | http://www.ncbi.nlm.nih.gov/pubmed/12419185 |
| PubMed | 12427738 | http://www.ncbi.nlm.nih.gov/pubmed/12427738 |
| PubMed | 12442274 | http://www.ncbi.nlm.nih.gov/pubmed/12442274 |
| PubMed | 12442275 | http://www.ncbi.nlm.nih.gov/pubmed/12442275 |
| PubMed | 12887909 | http://www.ncbi.nlm.nih.gov/pubmed/12887909 |
| PubMed | 12890688 | http://www.ncbi.nlm.nih.gov/pubmed/12890688 |
| PubMed | 12938098 | http://www.ncbi.nlm.nih.gov/pubmed/12938098 |
| PubMed | 14636569 | http://www.ncbi.nlm.nih.gov/pubmed/14636569 |
| PubMed | 14722926 | http://www.ncbi.nlm.nih.gov/pubmed/14722926 |
| PubMed | 14746861 | http://www.ncbi.nlm.nih.gov/pubmed/14746861 |
| PubMed | 14976165 | http://www.ncbi.nlm.nih.gov/pubmed/14976165 |
| PubMed | 14990569 | http://www.ncbi.nlm.nih.gov/pubmed/14990569 |
| PubMed | 15026808 | http://www.ncbi.nlm.nih.gov/pubmed/15026808 |
| PubMed | 15096610 | http://www.ncbi.nlm.nih.gov/pubmed/15096610 |
| PubMed | 15133502 | http://www.ncbi.nlm.nih.gov/pubmed/15133502 |
| PubMed | 15365993 | http://www.ncbi.nlm.nih.gov/pubmed/15365993 |
| PubMed | 15456891 | http://www.ncbi.nlm.nih.gov/pubmed/15456891 |
| PubMed | 15489334 | http://www.ncbi.nlm.nih.gov/pubmed/15489334 |
| PubMed | 15609993 | http://www.ncbi.nlm.nih.gov/pubmed/15609993 |
| PubMed | 16101277 | http://www.ncbi.nlm.nih.gov/pubmed/16101277 |
| PubMed | 16326698 | http://www.ncbi.nlm.nih.gov/pubmed/16326698 |
| PubMed | 16625196 | http://www.ncbi.nlm.nih.gov/pubmed/16625196 |
| PubMed | 16698035 | http://www.ncbi.nlm.nih.gov/pubmed/16698035 |
| PubMed | 16818604 | http://www.ncbi.nlm.nih.gov/pubmed/16818604 |
| PubMed | 16959974 | http://www.ncbi.nlm.nih.gov/pubmed/16959974 |
| PubMed | 17081983 | http://www.ncbi.nlm.nih.gov/pubmed/17081983 |
| PubMed | 17525332 | http://www.ncbi.nlm.nih.gov/pubmed/17525332 |
| PubMed | 17525340 | http://www.ncbi.nlm.nih.gov/pubmed/17525340 |
| PubMed | 17643121 | http://www.ncbi.nlm.nih.gov/pubmed/17643121 |
| PubMed | 17643122 | http://www.ncbi.nlm.nih.gov/pubmed/17643122 |
| PubMed | 17924331 | http://www.ncbi.nlm.nih.gov/pubmed/17924331 |
| PubMed | 18056443 | http://www.ncbi.nlm.nih.gov/pubmed/18056443 |
| PubMed | 18285836 | http://www.ncbi.nlm.nih.gov/pubmed/18285836 |
| PubMed | 18452305 | http://www.ncbi.nlm.nih.gov/pubmed/18452305 |
| PubMed | 18669648 | http://www.ncbi.nlm.nih.gov/pubmed/18669648 |
| PubMed | 18762988 | http://www.ncbi.nlm.nih.gov/pubmed/18762988 |
| PubMed | 19261746 | http://www.ncbi.nlm.nih.gov/pubmed/19261746 |
| PubMed | 19261748 | http://www.ncbi.nlm.nih.gov/pubmed/19261748 |
| PubMed | 19261749 | http://www.ncbi.nlm.nih.gov/pubmed/19261749 |
| PubMed | 19369211 | http://www.ncbi.nlm.nih.gov/pubmed/19369211 |
| PubMed | 19690332 | http://www.ncbi.nlm.nih.gov/pubmed/19690332 |
| PubMed | 20068231 | http://www.ncbi.nlm.nih.gov/pubmed/20068231 |
| PubMed | 20159462 | http://www.ncbi.nlm.nih.gov/pubmed/20159462 |
| PubMed | 20160719 | http://www.ncbi.nlm.nih.gov/pubmed/20160719 |
| PubMed | 20351172 | http://www.ncbi.nlm.nih.gov/pubmed/20351172 |
| PubMed | 20364141 | http://www.ncbi.nlm.nih.gov/pubmed/20364141 |
| PubMed | 20513136 | http://www.ncbi.nlm.nih.gov/pubmed/20513136 |
| PubMed | 21144835 | http://www.ncbi.nlm.nih.gov/pubmed/21144835 |
| PubMed | 21406692 | http://www.ncbi.nlm.nih.gov/pubmed/21406692 |
| PubMed | 21473589 | http://www.ncbi.nlm.nih.gov/pubmed/21473589 |
| PubMed | 21673012 | http://www.ncbi.nlm.nih.gov/pubmed/21673012 |
| PubMed | 22814378 | http://www.ncbi.nlm.nih.gov/pubmed/22814378 |
| PubMed | 23186163 | http://www.ncbi.nlm.nih.gov/pubmed/23186163 |
| PubMed | 23867111 | http://www.ncbi.nlm.nih.gov/pubmed/23867111 |
| PubMed | 24316840 | http://www.ncbi.nlm.nih.gov/pubmed/24316840 |
| PubMed | 25218447 | http://www.ncbi.nlm.nih.gov/pubmed/25218447 |
| PubMed | 25755297 | http://www.ncbi.nlm.nih.gov/pubmed/25755297 |
| PubMed | 26778126 | http://www.ncbi.nlm.nih.gov/pubmed/26778126 |
| PubMed | 26807646 | http://www.ncbi.nlm.nih.gov/pubmed/26807646 |
| PubMed | 7545954 | http://www.ncbi.nlm.nih.gov/pubmed/7545954 |
| PubMed | 7894491 | http://www.ncbi.nlm.nih.gov/pubmed/7894491 |
| PubMed | 7894493 | http://www.ncbi.nlm.nih.gov/pubmed/7894493 |
| PubMed | 7939630 | http://www.ncbi.nlm.nih.gov/pubmed/7939630 |
| PubMed | 8554067 | http://www.ncbi.nlm.nih.gov/pubmed/8554067 |
| PubMed | 8723683 | http://www.ncbi.nlm.nih.gov/pubmed/8723683 |
| PubMed | 8776600 | http://www.ncbi.nlm.nih.gov/pubmed/8776600 |
| PubMed | 8807330 | http://www.ncbi.nlm.nih.gov/pubmed/8807330 |
| PubMed | 8938427 | http://www.ncbi.nlm.nih.gov/pubmed/8938427 |
| PubMed | 8968716 | http://www.ncbi.nlm.nih.gov/pubmed/8968716 |
| PubMed | 8972225 | http://www.ncbi.nlm.nih.gov/pubmed/8972225 |
| PubMed | 9010228 | http://www.ncbi.nlm.nih.gov/pubmed/9010228 |
| PubMed | 9482581 | http://www.ncbi.nlm.nih.gov/pubmed/9482581 |
| PubMed | 9528852 | http://www.ncbi.nlm.nih.gov/pubmed/9528852 |
| PubMed | 9609997 | http://www.ncbi.nlm.nih.gov/pubmed/9609997 |
| PubMed | 9760198 | http://www.ncbi.nlm.nih.gov/pubmed/9760198 |
| PubMed | 9811458 | http://www.ncbi.nlm.nih.gov/pubmed/9811458 |
| Reactome | R-HSA-1221632 | http://www.reactome.org/cgi-bin/eventbrowser_st_id?ST_ID=R-HSA-1221632 |
| Reactome | R-HSA-3108214 | http://www.reactome.org/cgi-bin/eventbrowser_st_id?ST_ID=R-HSA-3108214 |
| Reactome | R-HSA-5685938 | http://www.reactome.org/cgi-bin/eventbrowser_st_id?ST_ID=R-HSA-5685938 |
| Reactome | R-HSA-5685942 | http://www.reactome.org/cgi-bin/eventbrowser_st_id?ST_ID=R-HSA-5685942 |
| Reactome | R-HSA-5689901 | http://www.reactome.org/cgi-bin/eventbrowser_st_id?ST_ID=R-HSA-5689901 |
| Reactome | R-HSA-5693554 | http://www.reactome.org/cgi-bin/eventbrowser_st_id?ST_ID=R-HSA-5693554 |
| Reactome | R-HSA-5693565 | http://www.reactome.org/cgi-bin/eventbrowser_st_id?ST_ID=R-HSA-5693565 |
| Reactome | R-HSA-5693568 | http://www.reactome.org/cgi-bin/eventbrowser_st_id?ST_ID=R-HSA-5693568 |
| Reactome | R-HSA-5693571 | http://www.reactome.org/cgi-bin/eventbrowser_st_id?ST_ID=R-HSA-5693571 |
| Reactome | R-HSA-5693579 | http://www.reactome.org/cgi-bin/eventbrowser_st_id?ST_ID=R-HSA-5693579 |
| Reactome | R-HSA-5693607 | http://www.reactome.org/cgi-bin/eventbrowser_st_id?ST_ID=R-HSA-5693607 |
| Reactome | R-HSA-5693616 | http://www.reactome.org/cgi-bin/eventbrowser_st_id?ST_ID=R-HSA-5693616 |
| Reactome | R-HSA-6796648 | http://www.reactome.org/cgi-bin/eventbrowser_st_id?ST_ID=R-HSA-6796648 |
| Reactome | R-HSA-6804756 | http://www.reactome.org/cgi-bin/eventbrowser_st_id?ST_ID=R-HSA-6804756 |
| Reactome | R-HSA-69473 | http://www.reactome.org/cgi-bin/eventbrowser_st_id?ST_ID=R-HSA-69473 |
| Reactome | R-HSA-912446 | http://www.reactome.org/cgi-bin/eventbrowser_st_id?ST_ID=R-HSA-912446 |
| RefSeq | NP_009225 | http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&id=NP_009225 |
| RefSeq | NP_009228 | http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&id=NP_009228 |
| RefSeq | NP_009229 | http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&id=NP_009229 |
| RefSeq | NP_009230 | http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&id=NP_009230 |
| RefSeq | NP_009231 | http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&id=NP_009231 |
| SMART | SM00184 | http://smart.embl.de/smart/do_annotation.pl?DOMAIN=SM00184 |
| SMART | SM00292 | http://smart.embl.de/smart/do_annotation.pl?DOMAIN=SM00292 |
| SMR | P38398 | http://swissmodel.expasy.org/repository/smr.php?sptr_ac=P38398 |
| STRING | 9606.ENSP00000418960 | http://string-db.org/newstring_cgi/show_network_section.pl?identifier=9606.ENSP00000418960&targetmode=cogs |
| SUPFAM | SSF52113 | http://supfam.org/SUPERFAMILY/cgi-bin/scop.cgi?ipid=SSF52113 |
| UCSC | uc002icq | http://genome.ucsc.edu/cgi-bin/hgGene?hgg_gene=uc002icq&org=rat |
| UCSC | uc010cyx | http://genome.ucsc.edu/cgi-bin/hgGene?hgg_gene=uc010cyx&org=rat |
| UniGene | Hs.194143 | http://www.ncbi.nlm.nih.gov/UniGene/clust.cgi?ORG=At&CID=Hs.194143 |
| UniProtKB-AC | P38398 | http://www.uniprot.org/uniprot/P38398 |
| UniProtKB | BRCA1_HUMAN | http://www.uniprot.org/uniprot/BRCA1_HUMAN |
| charge | swissprot:BRCA1_HUMAN | http://rest.g-language.org/emboss/charge/swissprot:BRCA1_HUMAN |
| eggNOG | ENOG410ITQ4 | http://eggnogapi.embl.de/nog_data/html/tree/ENOG410ITQ4 |
| eggNOG | ENOG4112BIH | http://eggnogapi.embl.de/nog_data/html/tree/ENOG4112BIH |
| epestfind | swissprot:BRCA1_HUMAN | http://rest.g-language.org/emboss/epestfind/swissprot:BRCA1_HUMAN |
| garnier | swissprot:BRCA1_HUMAN | http://rest.g-language.org/emboss/garnier/swissprot:BRCA1_HUMAN |
| helixturnhelix | swissprot:BRCA1_HUMAN | http://rest.g-language.org/emboss/helixturnhelix/swissprot:BRCA1_HUMAN |
| hmoment | swissprot:BRCA1_HUMAN | http://rest.g-language.org/emboss/hmoment/swissprot:BRCA1_HUMAN |
| iep | swissprot:BRCA1_HUMAN | http://rest.g-language.org/emboss/iep/swissprot:BRCA1_HUMAN |
| inforesidue | swissprot:BRCA1_HUMAN | http://rest.g-language.org/emboss/inforesidue/swissprot:BRCA1_HUMAN |
| neXtProt | NX_P38398 | http://www.nextprot.org/db/entry/NX_P38398 |
| octanol | swissprot:BRCA1_HUMAN | http://rest.g-language.org/emboss/octanol/swissprot:BRCA1_HUMAN |
| pepcoil | swissprot:BRCA1_HUMAN | http://rest.g-language.org/emboss/pepcoil/swissprot:BRCA1_HUMAN |
| pepdigest | swissprot:BRCA1_HUMAN | http://rest.g-language.org/emboss/pepdigest/swissprot:BRCA1_HUMAN |
| pepinfo | swissprot:BRCA1_HUMAN | http://rest.g-language.org/emboss/pepinfo/swissprot:BRCA1_HUMAN |
| pepnet | swissprot:BRCA1_HUMAN | http://rest.g-language.org/emboss/pepnet/swissprot:BRCA1_HUMAN |
| pepstats | swissprot:BRCA1_HUMAN | http://rest.g-language.org/emboss/pepstats/swissprot:BRCA1_HUMAN |
| pepwheel | swissprot:BRCA1_HUMAN | http://rest.g-language.org/emboss/pepwheel/swissprot:BRCA1_HUMAN |
| pepwindow | swissprot:BRCA1_HUMAN | http://rest.g-language.org/emboss/pepwindow/swissprot:BRCA1_HUMAN |
| sigcleave | swissprot:BRCA1_HUMAN | http://rest.g-language.org/emboss/sigcleave/swissprot:BRCA1_HUMAN |