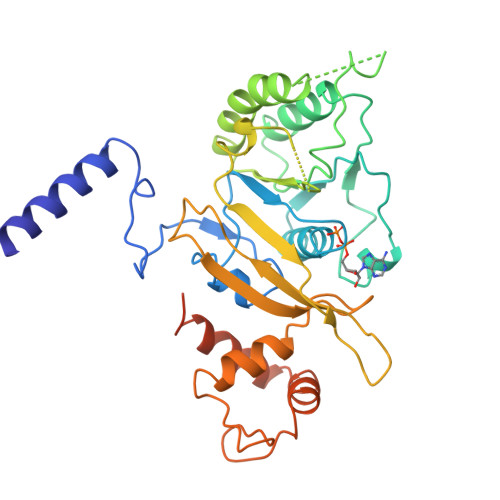
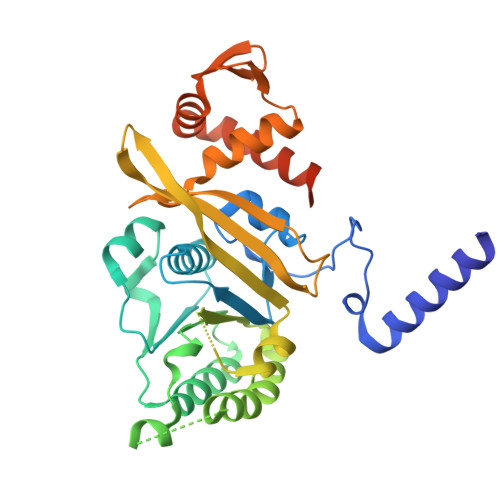
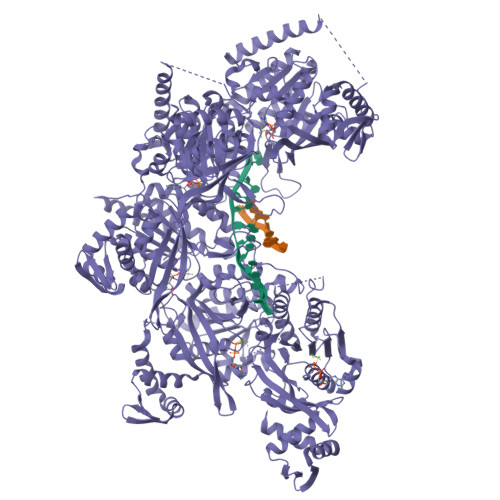
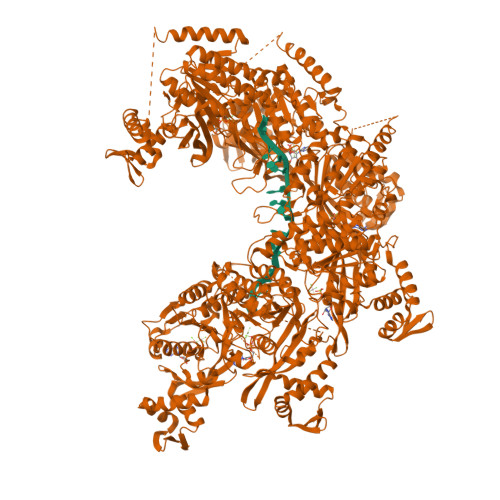
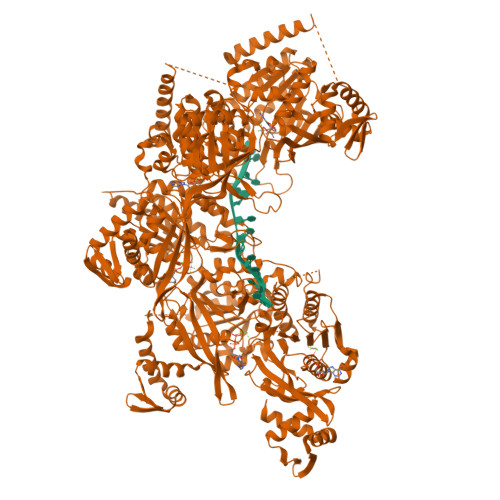
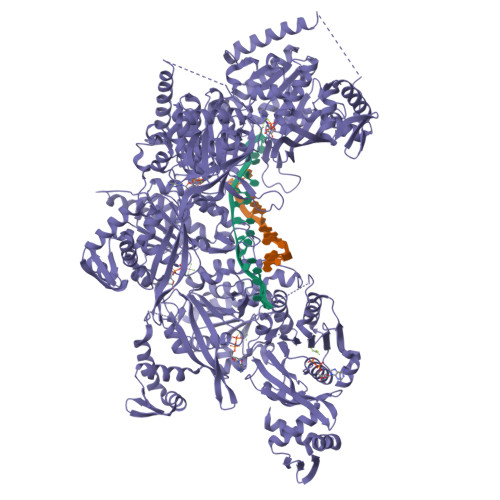
| # AltName | Recombinase A {ECO:0000255|HAMAP-Rule | MF_00268} |
| # BioGrid | 4259218 | 403 |
| # CDD | cd00983 | recA |
| # DISRUPTION PHENOTYPE | Triple mazE-mazF-recA mutant cells no longer undergo an apoptotic-like death upon DNA damage characterized by membrane depolarization (PubMed | 22412352). |
| # ENZYME REGULATION | The rate of DNA-strand exchange is stimulated by RadA. {ECO:0000269|PubMed | 26845522}. |
| # EcoGene | EG10823 | recA |
| # FUNCTION | RECA_ECOLI | Required for homologous recombination and the bypass of mutagenic DNA lesions by the SOS response. Catalyzes ATP-driven homologous pairing and strand exchange of DNA molecules necessary for DNA recombinational repair. Catalyzes the hydrolysis of ATP in the presence of single-stranded DNA, the ATP-dependent uptake of single-stranded DNA by duplex DNA, and the ATP-dependent hybridization of homologous single-stranded DNAs. The SOS response controls an apoptotic-like death (ALD) induced (in the absence of the mazE-mazF toxin-antitoxin module) in response to DNA damaging agents that is mediated by RecA and LexA (PubMed 22412352). {ECO 0000269|PubMed 22412352, ECO 0000269|PubMed 26845522, ECO 0000269|PubMed 7608206, ECO 0000269|PubMed 9230304}. |
| # GO_component | GO:0005829 | cytosol; IBA:GO_Central. |
| # GO_function | GO:0000150 | recombinase activity; IBA:GO_Central. |
| # GO_function | GO:0000400 | four-way junction DNA binding; IBA:GO_Central. |
| # GO_function | GO:0003684 | damaged DNA binding; IEA:UniProtKB-HAMAP. |
| # GO_function | GO:0003690 | double-stranded DNA binding; IBA:GO_Central. |
| # GO_function | GO:0003697 | single-stranded DNA binding; IDA:EcoliWiki. |
| # GO_function | GO:0005524 | ATP binding; IEA:UniProtKB-HAMAP. |
| # GO_function | GO:0008094 | DNA-dependent ATPase activity; IBA:GO_Central. |
| # GO_process | GO:0000730 | DNA recombinase assembly; IBA:GO_Central. |
| # GO_process | GO:0006310 | DNA recombination; IMP:CACAO. |
| # GO_process | GO:0006312 | mitotic recombination; IBA:GO_Central. |
| # GO_process | GO:0006974 | cellular response to DNA damage stimulus; IEP:EcoliWiki. |
| # GO_process | GO:0009432 | SOS response; IMP:CACAO. |
| # GO_process | GO:0010212 | response to ionizing radiation; IMP:EcoCyc. |
| # GO_process | GO:0042148 | strand invasion; IBA:GO_Central. |
| # GO_process | GO:0048870 | cell motility; IMP:EcoliWiki. |
| # GOslim_component | GO:0005829 | cytosol |
| # GOslim_function | GO:0003674 | molecular_function |
| # GOslim_function | GO:0003677 | DNA binding |
| # GOslim_function | GO:0016887 | ATPase activity |
| # GOslim_function | GO:0043167 | ion binding |
| # GOslim_process | GO:0006259 | DNA metabolic process |
| # GOslim_process | GO:0006950 | response to stress |
| # GOslim_process | GO:0008150 | biological_process |
| # GOslim_process | GO:0048870 | cell motility |
| # GOslim_process | GO:0065003 | macromolecular complex assembly |
| # Gene3D | 3.30.250.10 | -; 1. |
| # Gene3D | 3.40.50.300 | -; 1. |
| # HAMAP | MF_00268 | RecA |
| # INDUCTION | RECA_ECOLI | In response to low temperature. Sensitive to temperature through changes in the linking number of the DNA. 5.1- fold induced by hydroxyurea treatment (at protein level) (PubMed 20005847). mRNA levels are repressed in a mazE-mazF- mediated manner (PubMed 22412352). {ECO 0000269|PubMed 20005847, ECO 0000269|PubMed 22412352}. |
| # INTERACTION | RECA_ECOLI | P23367 mutL; NbExp=3; IntAct=EBI-370331, EBI-554913; |
| # IntAct | P0A7G6 | 28 |
| # InterPro | IPR003593 | AAA+_ATPase |
| # InterPro | IPR013765 | DNA_recomb/repair_RecA |
| # InterPro | IPR020584 | DNA_recomb/repair_RecA_CS |
| # InterPro | IPR020587 | RecA_monomer-monomer_interface |
| # InterPro | IPR020588 | RecA_ATP-bd |
| # InterPro | IPR023400 | RecA_C |
| # InterPro | IPR027417 | P-loop_NTPase |
| # KEGG_Brite | ko00001 | KEGG Orthology (KO) |
| # KEGG_Brite | ko03400 | DNA repair and recombination proteins |
| # KEGG_Pathway | ko03440 | Homologous recombination |
| # Organism | RECA_ECOLI | Escherichia coli (strain K12) |
| # PANTHER | PTHR22942:SF1 | PTHR22942:SF1 |
| # PATRIC | 32120796 | VBIEscCol129921_2790 |
| # PDB | 1AA3 | NMR; -; A=269-331 |
| # PDB | 1N03 | EM; 20.00 A; A/B/C/D/E/F/G=2-353 |
| # PDB | 1REA | X-ray; 2.70 A; A=2-353 |
| # PDB | 1U94 | X-ray; 1.90 A; A=2-353 |
| # PDB | 1U98 | X-ray; 2.00 A; A=2-353 |
| # PDB | 1U99 | X-ray; 2.60 A; A=2-353 |
| # PDB | 1XMS | X-ray; 2.10 A; A=2-353 |
| # PDB | 1XMV | X-ray; 1.90 A; A=2-353 |
| # PDB | 2REB | X-ray; 2.30 A; A=2-353 |
| # PDB | 2REC | EM; -; A/B/C/D/E/F=1-353 |
| # PDB | 3CMT | X-ray; 3.15 A; A/D=2-335 |
| # PDB | 3CMU | X-ray; 4.20 A; A=2-335 |
| # PDB | 3CMV | X-ray; 4.30 A; A/B/C/D/E/F/G/H=2-335 |
| # PDB | 3CMW | X-ray; 2.80 A; A/C=2-335 |
| # PDB | 3CMX | X-ray; 3.40 A; A/D=2-335 |
| # PDB | 4TWZ | X-ray; 2.80 A; A=2-353 |
| # PIR | G65049 | RQECA |
| # PRINTS | PR00142 | RECA |
| # PROSITE | PS00321 | RECA_1 |
| # PROSITE | PS50162 | RECA_2 |
| # PROSITE | PS50163 | RECA_3 |
| # Pfam | PF00154 | RecA |
| # Proteomes | UP000000318 | Chromosome |
| # Proteomes | UP000000625 | Chromosome |
| # RecName | Protein RecA {ECO:0000255|HAMAP-Rule | MF_00268} |
| # RefSeq | NP_417179 | NC_000913.3 |
| # RefSeq | WP_000963143 | NZ_LN832404.1 |
| # SIMILARITY | Belongs to the RecA family. {ECO:0000255|HAMAP- Rule | MF_00268}. |
| # SMART | SM00382 | AAA |
| # SUBCELLULAR LOCATION | RECA_ECOLI | Cytoplasm. |
| # SUBUNIT | RECA_ECOLI | Polymerizes non-specifically on ssDNA to form filaments; filament formation requires ATP or ATP-gamma-S. Interacts with and activates LexA leading to autocatalytic cleavage of LexA, which derepresses the SOS regulon and activates DNA repair. Interacts with the C-terminus of RecB, facilitating loading of RecA onto ssDNA at chi sites. Interaction is decreased by ATP. {ECO 0000269|PubMed 16483938, ECO 0000269|PubMed 1731246, ECO 0000269|PubMed 1731253}. |
| # SUPFAM | SSF52540 | SSF52540 |
| # SUPFAM | SSF54752 | SSF54752 |
| # TIGRFAMs | TIGR02012 | tigrfam_recA |
| # eggNOG | COG0468 | LUCA |
| # eggNOG | ENOG4105C68 | Bacteria |
| BLAST | swissprot:RECA_ECOLI | http://rest.g-language.org/emboss/kblast/swissprot:RECA_ECOLI |
| BioCyc | ECOL316407:JW2669-MONOMER | http://biocyc.org/getid?id=ECOL316407:JW2669-MONOMER |
| BioCyc | EcoCyc:EG10823-MONOMER | http://biocyc.org/getid?id=EcoCyc:EG10823-MONOMER |
| COG | COG0468 | http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=COG0468 |
| DIP | DIP-31832N | http://dip.doe-mbi.ucla.edu/dip/Browse.cgi?ID=DIP-31832N |
| DOI | 10.1002/elps.1150180805 | http://dx.doi.org/10.1002/elps.1150180805 |
| DOI | 10.1007/BF00633842 | http://dx.doi.org/10.1007/BF00633842 |
| DOI | 10.1016/S0092-8674(00)80315-3 | http://dx.doi.org/10.1016/S0092-8674(00)80315-3 |
| DOI | 10.1016/j.molcel.2006.01.007 | http://dx.doi.org/10.1016/j.molcel.2006.01.007 |
| DOI | 10.1016/j.molcel.2009.11.024 | http://dx.doi.org/10.1016/j.molcel.2009.11.024 |
| DOI | 10.1038/355318a0 | http://dx.doi.org/10.1038/355318a0 |
| DOI | 10.1038/355374a0 | http://dx.doi.org/10.1038/355374a0 |
| DOI | 10.1038/msb4100049 | http://dx.doi.org/10.1038/msb4100049 |
| DOI | 10.1038/nsb0297-101 | http://dx.doi.org/10.1038/nsb0297-101 |
| DOI | 10.1073/pnas.77.1.313 | http://dx.doi.org/10.1073/pnas.77.1.313 |
| DOI | 10.1073/pnas.77.5.2611 | http://dx.doi.org/10.1073/pnas.77.5.2611 |
| DOI | 10.1074/jbc.270.27.16360 | http://dx.doi.org/10.1074/jbc.270.27.16360 |
| DOI | 10.1093/dnares/4.2.91 | http://dx.doi.org/10.1093/dnares/4.2.91 |
| DOI | 10.1111/j.1432-1033.1995.0772m.x | http://dx.doi.org/10.1111/j.1432-1033.1995.0772m.x |
| DOI | 10.1111/j.1432-1033.1995.419_2.x | http://dx.doi.org/10.1111/j.1432-1033.1995.419_2.x |
| DOI | 10.1126/science.277.5331.1453 | http://dx.doi.org/10.1126/science.277.5331.1453 |
| DOI | 10.1371/journal.pbio.1001281 | http://dx.doi.org/10.1371/journal.pbio.1001281 |
| DOI | 10.7554/eLife.10807 | http://dx.doi.org/10.7554/eLife.10807 |
| EMBL | AP009048 | http://www.ebi.ac.uk/ena/data/view/AP009048 |
| EMBL | U00096 | http://www.ebi.ac.uk/ena/data/view/U00096 |
| EMBL | V00328 | http://www.ebi.ac.uk/ena/data/view/V00328 |
| EchoBASE | EB0816 | http://www.york.ac.uk/res/thomas/Gene.cfm?recordID=EB0816 |
| EcoGene | EG10823 | http://www.ecogene.org/geneInfo.php?eg_id=EG10823 |
| EnsemblBacteria | AAC75741 | http://www.ensemblgenomes.org/id/AAC75741 |
| EnsemblBacteria | AAC75741 | http://www.ensemblgenomes.org/id/AAC75741 |
| EnsemblBacteria | BAA16561 | http://www.ensemblgenomes.org/id/BAA16561 |
| EnsemblBacteria | BAA16561 | http://www.ensemblgenomes.org/id/BAA16561 |
| EnsemblBacteria | BAA16561 | http://www.ensemblgenomes.org/id/BAA16561 |
| EnsemblBacteria | b2699 | http://www.ensemblgenomes.org/id/b2699 |
| G-Links | GeneID:947170 | http://link.g-language.org/GeneID:947170/format=tsv |
| GO_component | GO:0005829 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0005829 |
| GO_function | GO:0000150 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0000150 |
| GO_function | GO:0000400 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0000400 |
| GO_function | GO:0003684 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0003684 |
| GO_function | GO:0003690 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0003690 |
| GO_function | GO:0003697 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0003697 |
| GO_function | GO:0005524 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0005524 |
| GO_function | GO:0008094 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0008094 |
| GO_process | GO:0000730 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0000730 |
| GO_process | GO:0006310 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0006310 |
| GO_process | GO:0006312 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0006312 |
| GO_process | GO:0006974 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0006974 |
| GO_process | GO:0009432 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0009432 |
| GO_process | GO:0010212 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0010212 |
| GO_process | GO:0042148 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0042148 |
| GO_process | GO:0048870 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0048870 |
| GOslim_component | GO:0005829 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0005829 |
| GOslim_function | GO:0003674 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0003674 |
| GOslim_function | GO:0003677 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0003677 |
| GOslim_function | GO:0016887 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0016887 |
| GOslim_function | GO:0043167 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0043167 |
| GOslim_process | GO:0006259 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0006259 |
| GOslim_process | GO:0006950 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0006950 |
| GOslim_process | GO:0008150 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0008150 |
| GOslim_process | GO:0048870 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0048870 |
| GOslim_process | GO:0065003 | http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0065003 |
| Gene3D | 3.30.250.10 | http://www.cathdb.info/version/latest/superfamily/3.30.250.10 |
| Gene3D | 3.40.50.300 | http://www.cathdb.info/version/latest/superfamily/3.40.50.300 |
| GeneID | 947170 | http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&term=947170 |
| HAMAP | MF_00268 | http://hamap.expasy.org/unirule/MF_00268 |
| HOGENOM | HOG000264120 | http://pbil.univ-lyon1.fr/cgi-bin/view-tree.pl?query=HOG000264120&db=HOGENOM6 |
| InParanoid | P0A7G6 | http://inparanoid.sbc.su.se/cgi-bin/gene_search.cgi?id=P0A7G6 |
| IntAct | P0A7G6 | http://www.ebi.ac.uk/intact/pages/interactions/interactions.xhtml?query=P0A7G6* |
| InterPro | IPR003593 | http://www.ebi.ac.uk/interpro/entry/IPR003593 |
| InterPro | IPR013765 | http://www.ebi.ac.uk/interpro/entry/IPR013765 |
| InterPro | IPR020584 | http://www.ebi.ac.uk/interpro/entry/IPR020584 |
| InterPro | IPR020587 | http://www.ebi.ac.uk/interpro/entry/IPR020587 |
| InterPro | IPR020588 | http://www.ebi.ac.uk/interpro/entry/IPR020588 |
| InterPro | IPR023400 | http://www.ebi.ac.uk/interpro/entry/IPR023400 |
| InterPro | IPR027417 | http://www.ebi.ac.uk/interpro/entry/IPR027417 |
| KEGG_Brite | ko00001 | http://www.genome.jp/dbget-bin/www_bget?ko00001 |
| KEGG_Brite | ko03400 | http://www.genome.jp/dbget-bin/www_bget?ko03400 |
| KEGG_Gene | ecj:JW2669 | http://www.genome.jp/dbget-bin/www_bget?ecj:JW2669 |
| KEGG_Gene | eco:b2699 | http://www.genome.jp/dbget-bin/www_bget?eco:b2699 |
| KEGG_Orthology | KO:K03553 | http://www.genome.jp/dbget-bin/www_bget?KO:K03553 |
| KEGG_Pathway | ko03440 | http://www.genome.jp/kegg-bin/show_pathway?ko03440 |
| MINT | MINT-1300726 | http://mint.bio.uniroma2.it/mint/search/search.do?queryType=protein&interactorAc=MINT-1300726 |
| OMA | PPFKEAH | http://omabrowser.org/cgi-bin/gateway.pl?f=DisplayGroup&p1=PPFKEAH |
| PANTHER | PTHR22942:SF1 | http://www.pantherdb.org/panther/family.do?clsAccession=PTHR22942:SF1 |
| PDB | 1AA3 | http://www.ebi.ac.uk/pdbe-srv/view/entry/1AA3 |
| PDB | 1N03 | http://www.ebi.ac.uk/pdbe-srv/view/entry/1N03 |
| PDB | 1REA | http://www.ebi.ac.uk/pdbe-srv/view/entry/1REA |
| PDB | 1U94 | http://www.ebi.ac.uk/pdbe-srv/view/entry/1U94 |
| PDB | 1U98 | http://www.ebi.ac.uk/pdbe-srv/view/entry/1U98 |
| PDB | 1U99 | http://www.ebi.ac.uk/pdbe-srv/view/entry/1U99 |
| PDB | 1XMS | http://www.ebi.ac.uk/pdbe-srv/view/entry/1XMS |
| PDB | 1XMV | http://www.ebi.ac.uk/pdbe-srv/view/entry/1XMV |
| PDB | 2REB | http://www.ebi.ac.uk/pdbe-srv/view/entry/2REB |
| PDB | 2REC | http://www.ebi.ac.uk/pdbe-srv/view/entry/2REC |
| PDB | 3CMT | http://www.ebi.ac.uk/pdbe-srv/view/entry/3CMT |
| PDB | 3CMU | http://www.ebi.ac.uk/pdbe-srv/view/entry/3CMU |
| PDB | 3CMV | http://www.ebi.ac.uk/pdbe-srv/view/entry/3CMV |
| PDB | 3CMW | http://www.ebi.ac.uk/pdbe-srv/view/entry/3CMW |
| PDB | 3CMX | http://www.ebi.ac.uk/pdbe-srv/view/entry/3CMX |
| PDB | 4TWZ | http://www.ebi.ac.uk/pdbe-srv/view/entry/4TWZ |
| PDBsum | 1AA3 | http://www.ebi.ac.uk/pdbsum/1AA3 |
| PDBsum | 1N03 | http://www.ebi.ac.uk/pdbsum/1N03 |
| PDBsum | 1REA | http://www.ebi.ac.uk/pdbsum/1REA |
| PDBsum | 1U94 | http://www.ebi.ac.uk/pdbsum/1U94 |
| PDBsum | 1U98 | http://www.ebi.ac.uk/pdbsum/1U98 |
| PDBsum | 1U99 | http://www.ebi.ac.uk/pdbsum/1U99 |
| PDBsum | 1XMS | http://www.ebi.ac.uk/pdbsum/1XMS |
| PDBsum | 1XMV | http://www.ebi.ac.uk/pdbsum/1XMV |
| PDBsum | 2REB | http://www.ebi.ac.uk/pdbsum/2REB |
| PDBsum | 2REC | http://www.ebi.ac.uk/pdbsum/2REC |
| PDBsum | 3CMT | http://www.ebi.ac.uk/pdbsum/3CMT |
| PDBsum | 3CMU | http://www.ebi.ac.uk/pdbsum/3CMU |
| PDBsum | 3CMV | http://www.ebi.ac.uk/pdbsum/3CMV |
| PDBsum | 3CMW | http://www.ebi.ac.uk/pdbsum/3CMW |
| PDBsum | 3CMX | http://www.ebi.ac.uk/pdbsum/3CMX |
| PDBsum | 4TWZ | http://www.ebi.ac.uk/pdbsum/4TWZ |
| PRINTS | PR00142 | http://umber.sbs.man.ac.uk/cgi-bin/dbbrowser/sprint/searchprintss.cgi?display_opts=Prints&category=None&queryform=false&prints_accn=PR00142 |
| PROSITE | PS00321 | http://prosite.expasy.org/cgi-bin/prosite/nicedoc.pl?PS00321 |
| PROSITE | PS50162 | http://prosite.expasy.org/cgi-bin/prosite/nicedoc.pl?PS50162 |
| PROSITE | PS50163 | http://prosite.expasy.org/cgi-bin/prosite/nicedoc.pl?PS50163 |
| PSORT-B | swissprot:RECA_ECOLI | http://rest.g-language.org/emboss/kpsortb/swissprot:RECA_ECOLI |
| PSORT2 | swissprot:RECA_ECOLI | http://rest.g-language.org/emboss/kpsort2/swissprot:RECA_ECOLI |
| PSORT | swissprot:RECA_ECOLI | http://rest.g-language.org/emboss/kpsort/swissprot:RECA_ECOLI |
| Pfam | PF00154 | http://pfam.xfam.org/family/PF00154 |
| Phobius | swissprot:RECA_ECOLI | http://rest.g-language.org/emboss/kphobius/swissprot:RECA_ECOLI |
| PhylomeDB | P0A7G6 | http://phylomedb.org/?seqid=P0A7G6 |
| ProteinModelPortal | P0A7G6 | http://www.proteinmodelportal.org/query/uniprot/P0A7G6 |
| PubMed | 16483938 | http://www.ncbi.nlm.nih.gov/pubmed/16483938 |
| PubMed | 16738553 | http://www.ncbi.nlm.nih.gov/pubmed/16738553 |
| PubMed | 1731246 | http://www.ncbi.nlm.nih.gov/pubmed/1731246 |
| PubMed | 1731253 | http://www.ncbi.nlm.nih.gov/pubmed/1731253 |
| PubMed | 20005847 | http://www.ncbi.nlm.nih.gov/pubmed/20005847 |
| PubMed | 22412352 | http://www.ncbi.nlm.nih.gov/pubmed/22412352 |
| PubMed | 2274037 | http://www.ncbi.nlm.nih.gov/pubmed/2274037 |
| PubMed | 26845522 | http://www.ncbi.nlm.nih.gov/pubmed/26845522 |
| PubMed | 6244554 | http://www.ncbi.nlm.nih.gov/pubmed/6244554 |
| PubMed | 6930655 | http://www.ncbi.nlm.nih.gov/pubmed/6930655 |
| PubMed | 7588783 | http://www.ncbi.nlm.nih.gov/pubmed/7588783 |
| PubMed | 7608206 | http://www.ncbi.nlm.nih.gov/pubmed/7608206 |
| PubMed | 7737176 | http://www.ncbi.nlm.nih.gov/pubmed/7737176 |
| PubMed | 9033586 | http://www.ncbi.nlm.nih.gov/pubmed/9033586 |
| PubMed | 9205837 | http://www.ncbi.nlm.nih.gov/pubmed/9205837 |
| PubMed | 9230304 | http://www.ncbi.nlm.nih.gov/pubmed/9230304 |
| PubMed | 9278503 | http://www.ncbi.nlm.nih.gov/pubmed/9278503 |
| PubMed | 9298644 | http://www.ncbi.nlm.nih.gov/pubmed/9298644 |
| RefSeq | NP_417179 | http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&id=NP_417179 |
| RefSeq | WP_000963143 | http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&id=WP_000963143 |
| SMART | SM00382 | http://smart.embl.de/smart/do_annotation.pl?DOMAIN=SM00382 |
| SMR | P0A7G6 | http://swissmodel.expasy.org/repository/smr.php?sptr_ac=P0A7G6 |
| STRING | 511145.b2699 | http://string-db.org/newstring_cgi/show_network_section.pl?identifier=511145.b2699&targetmode=cogs |
| STRING | COG0468 | http://string-db.org/newstring_cgi/show_network_section.pl?identifier=COG0468&targetmode=cogs |
| SUPFAM | SSF52540 | http://supfam.org/SUPERFAMILY/cgi-bin/scop.cgi?ipid=SSF52540 |
| SUPFAM | SSF54752 | http://supfam.org/SUPERFAMILY/cgi-bin/scop.cgi?ipid=SSF54752 |
| SWISS-2DPAGE | P0A7G6 | http://world-2dpage.expasy.org/swiss-2dpage/protein/ac=P0A7G6 |
| TIGRFAMs | TIGR02012 | http://cmr.jcvi.org/tigr-scripts/CMR/HmmReport.cgi?hmm_acc=TIGR02012 |
| UniProtKB-AC | P0A7G6 | http://www.uniprot.org/uniprot/P0A7G6 |
| UniProtKB | RECA_ECOLI | http://www.uniprot.org/uniprot/RECA_ECOLI |
| charge | swissprot:RECA_ECOLI | http://rest.g-language.org/emboss/charge/swissprot:RECA_ECOLI |
| eggNOG | COG0468 | http://eggnogapi.embl.de/nog_data/html/tree/COG0468 |
| eggNOG | ENOG4105C68 | http://eggnogapi.embl.de/nog_data/html/tree/ENOG4105C68 |
| epestfind | swissprot:RECA_ECOLI | http://rest.g-language.org/emboss/epestfind/swissprot:RECA_ECOLI |
| garnier | swissprot:RECA_ECOLI | http://rest.g-language.org/emboss/garnier/swissprot:RECA_ECOLI |
| helixturnhelix | swissprot:RECA_ECOLI | http://rest.g-language.org/emboss/helixturnhelix/swissprot:RECA_ECOLI |
| hmoment | swissprot:RECA_ECOLI | http://rest.g-language.org/emboss/hmoment/swissprot:RECA_ECOLI |
| iep | swissprot:RECA_ECOLI | http://rest.g-language.org/emboss/iep/swissprot:RECA_ECOLI |
| inforesidue | swissprot:RECA_ECOLI | http://rest.g-language.org/emboss/inforesidue/swissprot:RECA_ECOLI |
| octanol | swissprot:RECA_ECOLI | http://rest.g-language.org/emboss/octanol/swissprot:RECA_ECOLI |
| pepcoil | swissprot:RECA_ECOLI | http://rest.g-language.org/emboss/pepcoil/swissprot:RECA_ECOLI |
| pepdigest | swissprot:RECA_ECOLI | http://rest.g-language.org/emboss/pepdigest/swissprot:RECA_ECOLI |
| pepinfo | swissprot:RECA_ECOLI | http://rest.g-language.org/emboss/pepinfo/swissprot:RECA_ECOLI |
| pepnet | swissprot:RECA_ECOLI | http://rest.g-language.org/emboss/pepnet/swissprot:RECA_ECOLI |
| pepstats | swissprot:RECA_ECOLI | http://rest.g-language.org/emboss/pepstats/swissprot:RECA_ECOLI |
| pepwheel | swissprot:RECA_ECOLI | http://rest.g-language.org/emboss/pepwheel/swissprot:RECA_ECOLI |
| pepwindow | swissprot:RECA_ECOLI | http://rest.g-language.org/emboss/pepwindow/swissprot:RECA_ECOLI |
| sigcleave | swissprot:RECA_ECOLI | http://rest.g-language.org/emboss/sigcleave/swissprot:RECA_ECOLI |